

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0237.11
(176 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
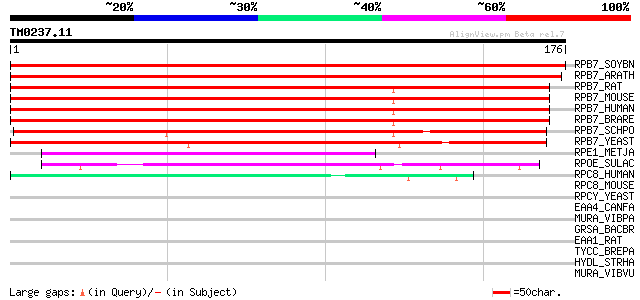
RPB7_SOYBN (P46279) DNA-directed RNA polymerase II 19 kDa polype... 352 2e-97
RPB7_ARATH (P38421) DNA-directed RNA polymerase II 19 kDa polype... 334 7e-92
RPB7_RAT (P62489) DNA-directed RNA polymerase II 19 kDa polypept... 209 3e-54
RPB7_MOUSE (P62488) DNA-directed RNA polymerase II 19 kDa polype... 209 3e-54
RPB7_HUMAN (P62487) DNA-directed RNA polymerase II 19 kDa polype... 209 3e-54
RPB7_BRARE (Q7ZW41) DNA-directed RNA polymerase II 19 kDa polype... 209 3e-54
RPB7_SCHPO (O14459) DNA-directed RNA polymerase II 19 kDa polype... 181 1e-45
RPB7_YEAST (P34087) DNA-directed RNA polymerase II 19 kDa polype... 134 1e-31
RPE1_METJA (Q57840) DNA-directed RNA polymerase subunit E' (EC 2... 59 8e-09
RPOE_SULAC (P39466) DNA-directed RNA polymerase subunit E (EC 2.... 50 3e-06
RPC8_HUMAN (Q9Y535) DNA-directed RNA polymerase III subunit 22.9... 43 3e-04
RPC8_MOUSE (Q9D2C6) DNA-directed RNA polymerase III subunit 22.9... 39 0.006
RPCY_YEAST (P35718) DNA-directed RNA polymerase III 25 kDa polyp... 33 0.26
EAA4_CANFA (Q9N1R2) Excitatory amino acid transporter 4 (Sodium-... 32 0.75
MURA_VIBPA (Q87LF4) UDP-N-acetylglucosamine 1-carboxyvinyltransf... 31 1.3
GRSA_BACBR (P14687) Gramicidin S synthetase I [Includes: ATP-dep... 31 1.7
EAA1_RAT (P24942) Excitatory amino acid transporter 1 (Sodium-de... 30 2.2
TYCC_BREPA (O30409) Tyrocidine synthetase III [Includes: ATP-dep... 30 2.9
HYDL_STRHA (Q05355) Putative polyketide hydroxylase (EC 1.14.13.-) 30 2.9
MURA_VIBVU (Q8DEB6) UDP-N-acetylglucosamine 1-carboxyvinyltransf... 30 3.7
>RPB7_SOYBN (P46279) DNA-directed RNA polymerase II 19 kDa
polypeptide (EC 2.7.7.6) (RNA polymerase II subunit 5)
Length = 176
Score = 352 bits (904), Expect = 2e-97
Identities = 171/176 (97%), Positives = 174/176 (98%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFFHIVLERNMQLHPRYFGRNLRDNLV+KLMKDVEGTCSGRHGFVVAVTGIEN+GKGLIR
Sbjct: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVSKLMKDVEGTCSGRHGFVVAVTGIENIGKGLIR 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ
Sbjct: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDPVAI 176
SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDP +
Sbjct: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDPATV 176
>RPB7_ARATH (P38421) DNA-directed RNA polymerase II 19 kDa
polypeptide (EC 2.7.7.6) (RNA polymerase II subunit 5)
Length = 176
Score = 334 bits (856), Expect = 7e-92
Identities = 159/175 (90%), Positives = 170/175 (96%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MFFHIVLERNMQLHPR+FGRNL++NLV+KLMKDVEGTCSGRHGFVVA+TGI+ +GKGLIR
Sbjct: 1 MFFHIVLERNMQLHPRFFGRNLKENLVSKLMKDVEGTCSGRHGFVVAITGIDTIGKGLIR 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
DGTGFVTFPVKYQCVVFRPFKGEILEAVVT+VNKMGFFAEAGPVQIFVS HLIPDDMEFQ
Sbjct: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTLVNKMGFFAEAGPVQIFVSKHLIPDDMEFQ 120
Query: 121 SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVINDPVA 175
+GDMPNYTTSDGSVKIQK+ EVRLKIIGTRVDAT IFC+GTIKDDFLGVINDP A
Sbjct: 121 AGDMPNYTTSDGSVKIQKECEVRLKIIGTRVDATAIFCVGTIKDDFLGVINDPAA 175
>RPB7_RAT (P62489) DNA-directed RNA polymerase II 19 kDa polypeptide
(EC 2.7.7.6) (RPB7)
Length = 172
Score = 209 bits (532), Expect = 3e-54
Identities = 95/172 (55%), Positives = 131/172 (75%), Gaps = 1/172 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE + LHPRYFG NL + + KL +VEGTC+G++GFV+AVT I+N+G G+I+
Sbjct: 1 MFYHISLEHEILLHPRYFGPNLLNTVKQKLFTEVEGTCTGKYGFVIAVTTIDNIGAGVIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G GFV +PVKY+ +VFRPFKGE+++AVVT VNK+G F E GP+ F+S H IP +MEF
Sbjct: 61 PGRGFVLYPVKYKAIVFRPFKGEVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFD 120
Query: 121 -SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVIN 171
+ + P Y T D + IQ+D E+RLKI+GTRVD +IF IG++ DD+LG+++
Sbjct: 121 PNSNPPCYKTMDEDIVIQQDDEIRLKIVGTRVDKNDIFAIGSLMDDYLGLVS 172
>RPB7_MOUSE (P62488) DNA-directed RNA polymerase II 19 kDa
polypeptide (EC 2.7.7.6) (RPB7)
Length = 172
Score = 209 bits (532), Expect = 3e-54
Identities = 95/172 (55%), Positives = 131/172 (75%), Gaps = 1/172 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE + LHPRYFG NL + + KL +VEGTC+G++GFV+AVT I+N+G G+I+
Sbjct: 1 MFYHISLEHEILLHPRYFGPNLLNTVKQKLFTEVEGTCTGKYGFVIAVTTIDNIGAGVIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G GFV +PVKY+ +VFRPFKGE+++AVVT VNK+G F E GP+ F+S H IP +MEF
Sbjct: 61 PGRGFVLYPVKYKAIVFRPFKGEVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFD 120
Query: 121 -SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVIN 171
+ + P Y T D + IQ+D E+RLKI+GTRVD +IF IG++ DD+LG+++
Sbjct: 121 PNSNPPCYKTMDEDIVIQQDDEIRLKIVGTRVDKNDIFAIGSLMDDYLGLVS 172
>RPB7_HUMAN (P62487) DNA-directed RNA polymerase II 19 kDa
polypeptide (EC 2.7.7.6) (RPB7)
Length = 172
Score = 209 bits (532), Expect = 3e-54
Identities = 95/172 (55%), Positives = 131/172 (75%), Gaps = 1/172 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE + LHPRYFG NL + + KL +VEGTC+G++GFV+AVT I+N+G G+I+
Sbjct: 1 MFYHISLEHEILLHPRYFGPNLLNTVKQKLFTEVEGTCTGKYGFVIAVTTIDNIGAGVIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G GFV +PVKY+ +VFRPFKGE+++AVVT VNK+G F E GP+ F+S H IP +MEF
Sbjct: 61 PGRGFVLYPVKYKAIVFRPFKGEVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFD 120
Query: 121 -SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVIN 171
+ + P Y T D + IQ+D E+RLKI+GTRVD +IF IG++ DD+LG+++
Sbjct: 121 PNSNPPCYKTMDEDIVIQQDDEIRLKIVGTRVDKNDIFAIGSLMDDYLGLVS 172
>RPB7_BRARE (Q7ZW41) DNA-directed RNA polymerase II 19 kDa
polypeptide (EC 2.7.7.6) (RPB7)
Length = 172
Score = 209 bits (532), Expect = 3e-54
Identities = 96/172 (55%), Positives = 131/172 (75%), Gaps = 1/172 (0%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF+HI LE + LHPRYFG NL + + KL +VEGTC+G++GFV+AVT I+N+G G+I+
Sbjct: 1 MFYHISLEHEILLHPRYFGPNLLNTVKQKLFTEVEGTCTGKYGFVIAVTTIDNIGAGVIQ 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G GFV +PVKY+ +VFRPFKGE+++AVVT VNK+G F E GP+ F+S H IP +MEF
Sbjct: 61 PGRGFVLYPVKYKAIVFRPFKGEVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFD 120
Query: 121 -SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVIN 171
+ + P Y T D V IQ+D E+RLKI+GTRVD +IF IG++ DD+LG+++
Sbjct: 121 PNSNPPCYKTVDEDVVIQQDDEIRLKIVGTRVDKNDIFAIGSLMDDYLGLVS 172
>RPB7_SCHPO (O14459) DNA-directed RNA polymerase II 19 kDa
polypeptide (EC 2.7.7.6)
Length = 172
Score = 181 bits (458), Expect = 1e-45
Identities = 89/172 (51%), Positives = 124/172 (71%), Gaps = 5/172 (2%)
Query: 2 FFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAV--TGIENVGKGLI 59
FF L + LHP YFG ++D L AKL+ DVEGTCSG++G+++ V + ++ KG +
Sbjct: 3 FFLKELSLTISLHPSYFGPRMQDYLKAKLLADVEGTCSGQYGYIICVLDSNTIDIDKGRV 62
Query: 60 RDGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEF 119
G GF F VKY+ V++RPF+GE+++A+VT VNKMGFFA GP+ +FVS+HL+P DM+F
Sbjct: 63 VPGQGFAEFEVKYRAVLWRPFRGEVVDAIVTTVNKMGFFANIGPLNVFVSSHLVPPDMKF 122
Query: 120 Q-SGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVI 170
+ + PNY+ D I+K S VRLKI+GTR DATEIF I T+K+D+LGV+
Sbjct: 123 DPTANPPNYSGED--QVIEKGSNVRLKIVGTRTDATEIFAIATMKEDYLGVL 172
>RPB7_YEAST (P34087) DNA-directed RNA polymerase II 19 kDa
polypeptide (EC 2.7.7.6) (B16)
Length = 171
Score = 134 bits (337), Expect = 1e-31
Identities = 67/173 (38%), Positives = 110/173 (62%), Gaps = 5/173 (2%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVG--KGL 58
MFF L N+ LHP +FG ++ L KL+++VEG+C+G+ G+++ V +N+ +G
Sbjct: 1 MFFIKDLSLNITLHPSFFGPRMKQYLKTKLLEEVEGSCTGKFGYILCVLDYDNIDIQRGR 60
Query: 59 IRDGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDME 118
I G F VKY+ VVF+PFKGE+++ V ++ GF + GP+++FV+ HL+P D+
Sbjct: 61 ILPTDGSAEFNVKYRAVVFKPFKGEVVDGTVVSCSQHGFEVQVGPMKVFVTKHLMPQDLT 120
Query: 119 FQSG-DMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEIFCIGTIKDDFLGVI 170
F +G + P+Y +S+ + I+ S +R+KI G + I IG+IK+D+LG I
Sbjct: 121 FNAGSNPPSYQSSEDVITIK--SRIRVKIEGCISQVSSIHAIGSIKEDYLGAI 171
>RPE1_METJA (Q57840) DNA-directed RNA polymerase subunit E' (EC
2.7.7.6)
Length = 187
Score = 58.5 bits (140), Expect = 8e-09
Identities = 30/106 (28%), Positives = 57/106 (53%)
Query: 11 MQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIRDGTGFVTFPV 70
+++ P FG++L++ + LM+ EG GFV+++ ++++G+G + G G PV
Sbjct: 11 VKVPPEEFGKDLKETVKKILMEKYEGRLDKDVGFVLSIVDVKDIGEGKVVHGDGSAYHPV 70
Query: 71 KYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDD 116
++ +V+ P E++E V V + G F GP+ + I DD
Sbjct: 71 VFETLVYIPEMYELIEGEVVDVVEFGSFVRLGPLDGLIHVSQIMDD 116
>RPOE_SULAC (P39466) DNA-directed RNA polymerase subunit E (EC
2.7.7.6)
Length = 248
Score = 49.7 bits (117), Expect = 3e-06
Identities = 46/169 (27%), Positives = 74/169 (43%), Gaps = 21/169 (12%)
Query: 11 MQLHPRYFGRN--------LRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIRDG 62
+++ P YFG++ LR KL+KD+ G V+ + + +G I G
Sbjct: 11 VRIPPEYFGQSVDEIAIKILRQEYQEKLIKDI--------GVVLGIVNAKASEEGFIIFG 62
Query: 63 TGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDD-MEFQS 121
G V++ +V+ P E++E V+ V+ G + GPV V I DD ++F S
Sbjct: 63 DGATYHEVEFDMLVYTPIIHEVIEGEVSQVDNYGVYVNMGPVDGLVHISQITDDNLKFDS 122
Query: 122 GDMPNYTTSDGSVK-IQKDSEVRLKIIGTRVDATEIFCIG-TIKDDFLG 168
+ S K I K VR II + + + I T+K +LG
Sbjct: 123 N--RGILIGEKSKKSITKGDRVRAMIISASMSSGRLPRIALTMKQPYLG 169
>RPC8_HUMAN (Q9Y535) DNA-directed RNA polymerase III subunit 22.9
kDa polypeptide (EC 2.7.7.6) (RPC8)
Length = 204
Score = 43.1 bits (100), Expect = 3e-04
Identities = 36/157 (22%), Positives = 63/157 (39%), Gaps = 14/157 (8%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF + + +++ P F R L D++ +L K + G + + I + +
Sbjct: 1 MFVLVEMVDTVRIPPWQFERKLNDSIAEELNKKLANKVVYNVGLCICLFDITKLEDAYVF 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G G V ++CVVF PF EIL + + G G F + LIP + Q
Sbjct: 61 PGDGASHTKVHFRCVVFHPFLDEILIGKIKGCSPEGVHVSLG----FFDDILIPPESLQQ 116
Query: 121 SGDMP--------NYTTSDGSVKIQKDS--EVRLKII 147
Y T +G+ + D+ E+R +++
Sbjct: 117 PAKFDEAEQVWVWEYETEEGAHDLYMDTGEEIRFRVV 153
>RPC8_MOUSE (Q9D2C6) DNA-directed RNA polymerase III subunit 22.9
kDa polypeptide (EC 2.7.7.6) (RPC8)
Length = 204
Score = 38.9 bits (89), Expect = 0.006
Identities = 35/157 (22%), Positives = 62/157 (39%), Gaps = 14/157 (8%)
Query: 1 MFFHIVLERNMQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIR 60
MF + + +++ P F R L D++ +L K + G + + I + +
Sbjct: 1 MFVLVEMVDTVRIPPWQFERKLNDSIAEELNKKLANKVVYNVGLCICLFDITKLEDAYVF 60
Query: 61 DGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQ 120
G G V ++ VVF PF EIL + + G G F + LIP + Q
Sbjct: 61 PGDGASHTKVHFRYVVFHPFLDEILIGKIKGCSPEGVHVSLG----FFDDILIPPESLQQ 116
Query: 121 SGDMP--------NYTTSDGSVKIQKDS--EVRLKII 147
Y T +G+ + D+ E+R +++
Sbjct: 117 PAKFDEAEQVWVWEYETEEGAHDLYMDTGEEIRFRVV 153
>RPCY_YEAST (P35718) DNA-directed RNA polymerase III 25 kDa
polypeptide (EC 2.7.7.6) (C25)
Length = 212
Score = 33.5 bits (75), Expect = 0.26
Identities = 30/134 (22%), Positives = 56/134 (41%), Gaps = 6/134 (4%)
Query: 11 MQLHPRYFGRNLRDNLVAKLMKDVEGTCSGRHGFVVAVTGIENVGKGLIRDGTGFVTFPV 70
+++ P F R+ + +L G + + + V +G ++ G G V
Sbjct: 11 VRIPPDQFHRDTISAITHQLNNKFANKIIPNVGLCITIYDLLTVEEGQLKPGDGSSYINV 70
Query: 71 KYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQIFVSNHLIPDDMEFQSGDMPNYTTS 130
++ VVF+PF GEI+ ++ G + + IF + IP +M F+ YT
Sbjct: 71 TFRAVVFKPFLGEIVTGWISKCTAEGI--KVSLLGIF-DDIFIPQNMLFEG---CYYTPE 124
Query: 131 DGSVKIQKDSEVRL 144
+ + D E +L
Sbjct: 125 ESAWIWPMDEETKL 138
>EAA4_CANFA (Q9N1R2) Excitatory amino acid transporter 4
(Sodium-dependent glutamate/aspartate transporter)
Length = 564
Score = 32.0 bits (71), Expect = 0.75
Identities = 22/82 (26%), Positives = 37/82 (44%)
Query: 47 AVTGIENVGKGLIRDGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQI 106
+ GI +G + G V VK++ V R F + EA++ MV + ++A G + +
Sbjct: 262 SANGINALGLVVFSVAFGLVIGGVKHKGRVLRDFFDSLNEAIMRMVGIIIWYAPVGILFL 321
Query: 107 FVSNHLIPDDMEFQSGDMPNYT 128
L +DM G + YT
Sbjct: 322 IAGKILEMEDMAVLGGQLGMYT 343
>MURA_VIBPA (Q87LF4) UDP-N-acetylglucosamine
1-carboxyvinyltransferase (EC 2.5.1.7) (Enoylpyruvate
transferase) (UDP-N-acetylglucosamine enolpyruvyl
transferase) (EPT)
Length = 418
Score = 31.2 bits (69), Expect = 1.3
Identities = 23/82 (28%), Positives = 33/82 (40%), Gaps = 7/82 (8%)
Query: 28 AKLMKDVEGTCSGRHGFVVAVTGIENVGKGL-------IRDGTGFVTFPVKYQCVVFRPF 80
A + + SG + + G+E +G G I GT V V VV R
Sbjct: 196 ADFLNKLGAKISGAGTDTITIEGVERLGGGKHSVVADRIETGTFLVAAAVSGGKVVCRNT 255
Query: 81 KGEILEAVVTMVNKMGFFAEAG 102
G +LEAV+ + + G E G
Sbjct: 256 NGHLLEAVLAKLEEAGALVETG 277
>GRSA_BACBR (P14687) Gramicidin S synthetase I [Includes:
ATP-dependent D-phenylalanine adenylase (D-PheA)
(D-phenylalanine activase); Phenylalanine racemase
[ATP-hydrolyzing] (EC 5.1.1.11)]
Length = 1098
Score = 30.8 bits (68), Expect = 1.7
Identities = 16/52 (30%), Positives = 30/52 (56%), Gaps = 1/52 (1%)
Query: 105 QIFVSNHLIPDDMEFQSGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEI 156
Q FV N +P + +++GD + SDG+++ + ++KI G RV+ E+
Sbjct: 395 QKFVDNPFVPGEKLYKTGDQARWL-SDGNIEYLGRIDNQVKIRGHRVELEEV 445
>EAA1_RAT (P24942) Excitatory amino acid transporter 1
(Sodium-dependent glutamate/aspartate transporter 1)
(Glial glutamate transporter) (GLAST) (GLAST-1)
Length = 543
Score = 30.4 bits (67), Expect = 2.2
Identities = 21/84 (25%), Positives = 38/84 (45%)
Query: 47 AVTGIENVGKGLIRDGTGFVTFPVKYQCVVFRPFKGEILEAVVTMVNKMGFFAEAGPVQI 106
+V G+ +G + GFV +K Q R F + EA++ +V + ++A G + +
Sbjct: 237 SVNGVNALGLVVFSMCFGFVIGNMKEQGQALREFFDSLNEAIMRLVAVIMWYAPLGILFL 296
Query: 107 FVSNHLIPDDMEFQSGDMPNYTTS 130
L +DM G + YT +
Sbjct: 297 IAGKILEMEDMGVIGGQLAMYTVT 320
>TYCC_BREPA (O30409) Tyrocidine synthetase III [Includes:
ATP-dependent asparagine adenylase (AsnA) (Asparagine
activase); ATP-dependent glutamine adenylase (GlnA)
(Glutamine activase); ATP-dependent tyrosine adenylase
(TyrA) (Tyrosine activase); ATP-depe
Length = 6486
Score = 30.0 bits (66), Expect = 2.9
Identities = 15/50 (30%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Query: 107 FVSNHLIPDDMEFQSGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEI 156
FV N P + +++GD+ + +DG+V+ ++ ++KI G R++ EI
Sbjct: 4978 FVPNPFAPGERLYRTGDLAKWR-ADGNVEFLGRNDHQVKIRGVRIELGEI 5026
Score = 29.6 bits (65), Expect = 3.7
Identities = 14/50 (28%), Positives = 31/50 (62%), Gaps = 1/50 (2%)
Query: 107 FVSNHLIPDDMEFQSGDMPNYTTSDGSVKIQKDSEVRLKIIGTRVDATEI 156
FV+N P + +++GD+ + T DG+++ ++ ++K+ G R++ EI
Sbjct: 6020 FVANPFRPGERMYKTGDLVKWRT-DGTIEYIGRADEQVKVRGYRIEIGEI 6068
>HYDL_STRHA (Q05355) Putative polyketide hydroxylase (EC 1.14.13.-)
Length = 555
Score = 30.0 bits (66), Expect = 2.9
Identities = 23/83 (27%), Positives = 39/83 (46%), Gaps = 11/83 (13%)
Query: 4 HIVLERNM--QLHPRYFGRNLRDNLVAKLMKDVEG------TCSGRHGFVVAVTGIENVG 55
H+++ER+ +HPR G N+R V + +G T +G HG + + + + G
Sbjct: 40 HMLVERHAGTSVHPRGRGNNVRTMEVYRAAGVEQGIRRAAATLAGNHGILQTPSLVGDEG 99
Query: 56 KGLIRD---GTGFVTFPVKYQCV 75
+ L+RD G G F C+
Sbjct: 100 EWLLRDIDPGGGLARFSPSSWCL 122
>MURA_VIBVU (Q8DEB6) UDP-N-acetylglucosamine
1-carboxyvinyltransferase (EC 2.5.1.7) (Enoylpyruvate
transferase) (UDP-N-acetylglucosamine enolpyruvyl
transferase) (EPT)
Length = 421
Score = 29.6 bits (65), Expect = 3.7
Identities = 22/82 (26%), Positives = 33/82 (39%), Gaps = 7/82 (8%)
Query: 28 AKLMKDVEGTCSGRHGFVVAVTGIENVGKGL-------IRDGTGFVTFPVKYQCVVFRPF 80
AK + + SG + + G+E +G G I GT V V VV
Sbjct: 196 AKFLNTLGAKISGAGTDTITIEGVERLGGGKHAVVADRIETGTFLVAAAVSGGKVVCHNT 255
Query: 81 KGEILEAVVTMVNKMGFFAEAG 102
+ +LEAV+ + + G E G
Sbjct: 256 QAHLLEAVLAKLEEAGALVETG 277
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.144 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,948,288
Number of Sequences: 164201
Number of extensions: 836943
Number of successful extensions: 2104
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 2071
Number of HSP's gapped (non-prelim): 35
length of query: 176
length of database: 59,974,054
effective HSP length: 103
effective length of query: 73
effective length of database: 43,061,351
effective search space: 3143478623
effective search space used: 3143478623
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0237.11


