

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0234.16
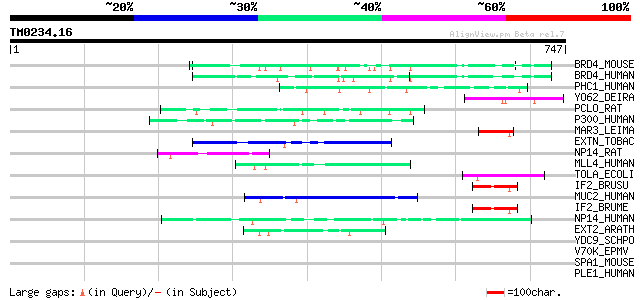
(747 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BRD4_MOUSE (Q9ESU6) Bromodomain-containing protein 4 (Mitotic ch... 63 2e-09
BRD4_HUMAN (O60885) Bromodomain-containing protein 4 (HUNK1 prot... 58 8e-08
PHC1_HUMAN (P78364) Polyhomeotic-like protein 1 (Early developme... 49 4e-05
YO62_DEIRA (Q9RRM6) Hypothetical UPF0144 protein DR2462 49 5e-05
PCLO_RAT (Q9JKS6) Piccolo protein (Multidomain presynaptic cytom... 48 1e-04
P300_HUMAN (Q09472) E1A-associated protein p300 (EC 2.3.1.48) 47 2e-04
MAR3_LEIMA (P14700) Membrane antigen containing repeating peptid... 47 2e-04
EXTN_TOBAC (P13983) Extensin precursor (Cell wall hydroxyproline... 47 2e-04
NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 k... 46 3e-04
MLL4_HUMAN (Q9UMN6) Myeloid/lymphoid or mixed-lineage leukemia p... 46 3e-04
TOLA_ECOLI (P19934) TolA protein 46 4e-04
IF2_BRUSU (Q8FXT2) Translation initiation factor IF-2 46 4e-04
MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2) 45 5e-04
IF2_BRUME (Q8YEB3) Translation initiation factor IF-2 45 5e-04
NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130... 45 7e-04
EXT2_ARATH (Q9M1G9) Extensin 2 precursor (AtExt2) (Cell wall hyd... 45 7e-04
YDC9_SCHPO (Q10172) Hypothetical protein C25G10.09c in chromosome I 44 0.001
V70K_EPMV (P20129) 70 kDa protein 44 0.001
SPA1_MOUSE (P46062) Signal-induced proliferation associated prot... 44 0.001
PLE1_HUMAN (Q15149) Plectin 1 (PLTN) (PCN) (Hemidesmosomal prote... 44 0.001
>BRD4_MOUSE (Q9ESU6) Bromodomain-containing protein 4 (Mitotic
chromosome-associated protein) (MCAP)
Length = 1400
Score = 63.2 bits (152), Expect = 2e-09
Identities = 112/529 (21%), Positives = 189/529 (35%), Gaps = 108/529 (20%)
Query: 243 DDGPPSPKKQKKQVRIVVKPT-RVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNL 301
D+ PP+P Q+++ ++ +V+P ++ V+ ++ +V
Sbjct: 917 DEEPPAPPLTSMQMQLYLQQLQKVQPPTPLLPSVKVQSQPPPPLPPPPHPSVQQQQ---- 972
Query: 302 FDALPISALLKQSSNPLTPIPESQPAAQTTSPP---HSPRSSFFQ------PSPNEAPLW 352
L Q P P P+ P Q PP H P F P P + P
Sbjct: 973 --------LQPQPPPPPPPQPQPPPQQQHQPPPRPVHLPSMPFSAHIQQPPPPPGQQPTH 1024
Query: 353 NNFRTNPPDP-------LIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLL-- 403
PP P +IQ HPSP H++ ++ P P+ ++ Q+ L
Sbjct: 1025 PPPGQQPPPPQPAKPQQVIQHHPSPRHHKSDPYSAGHLREAPSPLMIHSPQMPQFQSLTH 1084
Query: 404 ---PPITLNPLEN-SWRSEKEKVSTLEEYYLTCP--SPRRYP----GPRPERLVDPD--- 450
P + P + R+E + + CP SP P RP +V P
Sbjct: 1085 QSPPQQNVQPKKQVKGRAEPQPPGPVMGQGQGCPPASPAAVPMLSQELRPPSVVQPQPLV 1144
Query: 451 ---EPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPH 507
E + +P+ ++P + ++P P P PE ++++P L
Sbjct: 1145 VVKEEKIHSPIIRSEPFSTSLRPEP----PKHPE-----------NIKAPVHL------- 1182
Query: 508 LGASEPHVQTCDIGSPQGASEAHSSNHPASP-------ETNLSIIPYTHLRPTSLSECIN 560
P ++ DIG P S+ P +P E + P L+ ++ +
Sbjct: 1183 --PQRPEMKPVDIGRPVIRPPEQSAPPPGAPDKDKQKQEPKTPVAPKKDLKIKNMGSWAS 1240
Query: 561 IFHQEASLMLCNVQGQTDLSENADYVDEEWHSLSTWLVAQVPVIMQLLHAEGSQRIEAAK 620
+ + + + +D E+ E L AQ HAE E +
Sbjct: 1241 LVQKHPTTPSSTAKSSSDSFEHFRRAAREKEEREKALKAQAE------HAEK----EKER 1290
Query: 621 QRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAERLEAEAEAR 680
R R + +++ ++ A EEARR+QEQ + Q +Q R E ++ + +A A
Sbjct: 1291 LRQERMRSREDEDALEQARRAHEEARRRQEQQQ--------QQQQQRQEQQQQQQQAAA- 1341
Query: 681 RLAPVVFTPVASASTPANQAAQNVPSSSTHSSSSRLDTVEQRLNTHESM 729
VA+AS P Q++Q P S EQ E+M
Sbjct: 1342 ---------VAAASAPQAQSSQ--PQSMLDQQRELARKREQERRRREAM 1379
Score = 55.8 bits (133), Expect = 4e-07
Identities = 108/481 (22%), Positives = 163/481 (33%), Gaps = 95/481 (19%)
Query: 246 PPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDAL 305
PP+ Q ++ +P V+P ++ E PA S L
Sbjct: 889 PPAVSPALAQPPLLPQPPMVQPPQVLLEDEEPPAPPLTSMQMQ----------------L 932
Query: 306 PISALLK-QSSNPLTPIP--ESQPAAQTTSPPH-SPRSSFFQPSPNEAPLWNNFRTNPPD 361
+ L K Q PL P +SQP PPH S + QP P PP
Sbjct: 933 YLQQLQKVQPPTPLLPSVKVQSQPPPPLPPPPHPSVQQQQLQPQP------------PPP 980
Query: 362 PLIQPHPSPSKHQNVWLPELLIQTLPQPIHL--YPSQLMLLTLLPPITLNPLEN--SWRS 417
P QP P P + Q P+P+HL P + PP P +
Sbjct: 981 PPPQPQPPPQQQH---------QPPPRPVHLPSMPFSAHIQQPPPPPGQQPTHPPPGQQP 1031
Query: 418 EKEKVSTLEEYYLTCPSPRRYPG-----------PRPERLVDPDEPILANPLHEADPLAQ 466
+ + ++ PSPR + P P + P P + H++ P Q
Sbjct: 1032 PPPQPAKPQQVIQHHPSPRHHKSDPYSAGHLREAPSPLMIHSPQMPQFQSLTHQSPP-QQ 1090
Query: 467 QVQPAPHQQQPVQPEPE----------PEQSVS---------NHSSVRSPNPLVATSEPH 507
VQP + +P+P P S + SV P PLV E
Sbjct: 1091 NVQPKKQVKGRAEPQPPGPVMGQGQGCPPASPAAVPMLSQELRPPSVVQPQPLVVVKEEK 1150
Query: 508 LGA----SEPHVQTCDIGSPQGASEAHSSNH-PASPETNLSIIPYTHLRPTSLSECINIF 562
+ + SEP + P+ + H P PE I +RP S
Sbjct: 1151 IHSPIIRSEPFSTSLRPEPPKHPENIKAPVHLPQRPEMKPVDIGRPVIRPPEQSA----- 1205
Query: 563 HQEASLMLCNVQGQTDLSENADYVDEEWHSLSTW--LVAQVPVIMQLLHAEGSQRIEAAK 620
+ Q Q + A D + ++ +W LV + P S E +
Sbjct: 1206 -PPPGAPDKDKQKQEPKTPVAPKKDLKIKNMGSWASLVQKHPTTPSSTAKSSSDSFEHFR 1264
Query: 621 QRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAERLEAEAEAR 680
+ A E+E+R+K L+A E K+++ R+ + +DE A +A R EA R
Sbjct: 1265 R------AAREKEEREKALKAQAEHAEKEKERLRQERMRSREDEDALEQARRAHEEARRR 1318
Query: 681 R 681
+
Sbjct: 1319 Q 1319
Score = 35.8 bits (81), Expect = 0.42
Identities = 86/422 (20%), Positives = 144/422 (33%), Gaps = 68/422 (16%)
Query: 186 PAPEFDPPLTKKVQRKMILEDSSEESDVPLNKGKKAG------QSKRKHDETSKPDDGDD 239
P P ++ K + E D+ G+K G QS+ + S PD+ +
Sbjct: 596 PPPTYESEEEDKCKPMSYEEKRQLSLDINKLPGEKLGRVVHIIQSREPSLKNSNPDEIEI 655
Query: 240 GDNDDGPPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARV---TRSSAQSSKSAVASD 296
P + ++ ++ V ++ R +P AE + + +++ + S ++S+ + +SD
Sbjct: 656 DFETLKPSTLRELERYVTSCLRKKR-KPQAEKVDVIAGSSKMKGFSSSESESTSESSSSD 714
Query: 297 DDLNLFDALPISALL---------------KQSSNPLTPIPESQ-PAAQTTSPPHSPRSS 340
+ + + P S Q P+P+ P Q PP P+
Sbjct: 715 SEDSETEMAPKSKKKGHTGRDQKKHHHHHHPQMQPAPAPVPQQPPPPPQQPPPPPPPQQQ 774
Query: 341 FFQPSPNEAP------LWNNFRTNPPD------PLIQPHPSPSKHQNVWLPELLIQTLPQ 388
QP P P +++PP P+++P S + I LPQ
Sbjct: 775 QQQPPPPPPPPSMPQQTAPAMKSSPPPFITAQVPVLEPQLPGSVFDPIGHFTQPILHLPQ 834
Query: 389 ----------PIHLYPSQL-MLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRR 437
P H P L + PP N L + + L P P R
Sbjct: 835 PELPPHLPQPPEHSTPPHLNQHAVVSPPALHNALPQQPSRPSNRAAALP------PKPTR 888
Query: 438 YPGPRP----------ERLVDPDEPILANPLHEADPL-AQQVQPAPHQQQPVQPEPEPEQ 486
P P +V P + +L + A PL + Q+Q Q Q VQP
Sbjct: 889 PPAVSPALAQPPLLPQPPMVQPPQVLLEDEEPPAPPLTSMQMQLYLQQLQKVQPPTPLLP 948
Query: 487 SVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPASPETNLSIIP 546
SV S + P PL P + + Q PQ + P +L +P
Sbjct: 949 SVKVQS--QPPPPLPPPPHPSVQQQQLQPQPPPPPPPQPQPPPQQQHQPPPRPVHLPSMP 1006
Query: 547 YT 548
++
Sbjct: 1007 FS 1008
Score = 32.0 bits (71), Expect = 6.1
Identities = 34/127 (26%), Positives = 47/127 (36%), Gaps = 13/127 (10%)
Query: 318 LTPIPESQPAAQTTSPPH---SPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQ 374
+TP +QP T PP +P QP P AP+ +++PP P P +K
Sbjct: 229 VTPDLIAQPPVMTMVPPQPLQTPSPVPPQPPPPPAPVPQPVQSHPPIIATTPQPVKTKKG 288
Query: 375 NVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPS 434
+ T PIH PS L L P S R K + P
Sbjct: 289 VKRKADTTTPTTIDPIHEPPS---LAPEPKTAKLGPRRESSRPVKPPKKDV-------PD 338
Query: 435 PRRYPGP 441
+++PGP
Sbjct: 339 SQQHPGP 345
>BRD4_HUMAN (O60885) Bromodomain-containing protein 4 (HUNK1 protein)
Length = 1362
Score = 58.2 bits (139), Expect = 8e-08
Identities = 112/543 (20%), Positives = 174/543 (31%), Gaps = 147/543 (27%)
Query: 246 PPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDAL 305
PP+ Q ++ +P +P ++ E PA S L
Sbjct: 887 PPAVSPALTQTPLLPQPPMAQPPQVLLEDEEPPAPPLTSMQMQ----------------L 930
Query: 306 PISALLK-QSSNPLTPIP--ESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDP 362
+ L K Q PL P +SQP PPH Q P PP P
Sbjct: 931 YLQQLQKVQPPTPLLPSVKVQSQPPPPLPPPPHPSVQQQLQQQP------------PPPP 978
Query: 363 LIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKV 422
QP P P + Q P+P+HL P Q PP P +
Sbjct: 979 PPQPQPPPQQQH---------QPPPRPVHLQPMQFSTHIQQPP----PPQGQQPPH---- 1021
Query: 423 STLEEYYLTCPSPRRYPGP----RPERLVD--------PDEPILANPLHEA-------DP 463
P P + P P +P++++ +P L EA P
Sbjct: 1022 ----------PPPGQQPPPPQPAKPQQVIQHHHSPRHHKSDPYSTGHLREAPSPLMIHSP 1071
Query: 464 LAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGA------------- 510
Q Q HQ P Q +P++ +SV P PLV E + +
Sbjct: 1072 QMSQFQSLTHQSPP-QQNVQPKKQELRAASVVQPQPLVVVKEEKIHSPIIRSEPFSPSLR 1130
Query: 511 -----------------SEPHVQTCDIGSPQGASEAHSSNHPASP-------ETNLSIIP 546
P ++ D+G P ++ P +P E + P
Sbjct: 1131 PEPPKHPESIKAPVHLPQRPEMKPVDVGRPVIRPPEQNAPPPGAPDKDKQKQEPKTPVAP 1190
Query: 547 YTHLRPTSLSECINIFHQEASLMLCNVQGQTDLSENADYVDEEWHSLSTWLVAQVPVIMQ 606
L+ ++ ++ + + + +D E E L AQ
Sbjct: 1191 KKDLKIKNMGSWASLVQKHPTTPSSTAKSSSDSFEQFRRAAREKEEREKALKAQAE---- 1246
Query: 607 LLHAEGSQRIEAAKQRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQA 666
HAE E + R R + +++ ++ A EEARR+QEQ Q +Q
Sbjct: 1247 --HAEK----EKERLRQERMRSREDEDALEQARRAHEEARRRQEQ----------QQQQR 1290
Query: 667 RLEAERLEAEAEARRLAPVVFTPVASASTPANQAAQNVPSSSTHSSSSRLDTVEQRLNTH 726
+ + ++ + +A A VA+A+TP Q++Q P S EQ
Sbjct: 1291 QEQQQQQQQQAAA----------VAAAATPQAQSSQ--PQSMLDQQRELARKREQERRRR 1338
Query: 727 ESM 729
E+M
Sbjct: 1339 EAM 1341
Score = 49.7 bits (117), Expect = 3e-05
Identities = 74/307 (24%), Positives = 113/307 (36%), Gaps = 34/307 (11%)
Query: 246 PPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDAL 305
PP P++Q++ P+ + AA ++ P T+ + + D + F
Sbjct: 767 PPPPQQQQQPPPPPPPPSMPQQAAPAMKSSPPPFIATQVPVLEPQLPGSVFDPIGHFTQ- 825
Query: 306 PISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSP--NEAPLWNNFRTN----- 358
PI L + P P P ++PPH + + P N P + +N
Sbjct: 826 PILHLPQPELPPHLPQPPEH-----STPPHLNQHAVVSPPALHNALPQQPSRPSNRAAAL 880
Query: 359 PPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLEN-SWRS 417
PP P P SP+ Q LPQP P Q++L PP PL + +
Sbjct: 881 PPKPARPPAVSPALTQT--------PLLPQPPMAQPPQVLLEDEEPPAP--PLTSMQMQL 930
Query: 418 EKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQP 477
+++ ++ PS + P P L P P + L + P QP P QQ
Sbjct: 931 YLQQLQKVQPPTPLLPSVKVQSQPPPP-LPPPPHPSVQQQLQQQPPPPPPPQPQPPPQQQ 989
Query: 478 VQPEPEPE--QSVSNHSSVRSPNPLVATSEPH--LGASEPHVQTCDIGSPQGASEAHSS- 532
QP P P Q + + ++ P P PH G P Q PQ + H S
Sbjct: 990 HQPPPRPVHLQPMQFSTHIQQPPPPQGQQPPHPPPGQQPPPPQP---AKPQQVIQHHHSP 1046
Query: 533 -NHPASP 538
+H + P
Sbjct: 1047 RHHKSDP 1053
Score = 35.4 bits (80), Expect = 0.55
Identities = 79/372 (21%), Positives = 127/372 (33%), Gaps = 65/372 (17%)
Query: 186 PAPEFDPPLTKKVQRKMILEDSSEESDVPLNKGKKAG------QSKRKHDETSKPDDGDD 239
P P ++ K + E D+ G+K G QS+ + S PD+ +
Sbjct: 595 PPPTYESEEEDKCKPMSYEEKRQLSLDINKLPGEKLGRVVHIIQSREPSLKNSNPDEIEI 654
Query: 240 GDNDDGPPSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARV---TRSSAQSSKSAVASD 296
P + ++ ++ V ++ R +P AE + + +++ + S ++SS + +SD
Sbjct: 655 DFETLKPSTLRELERYVTSCLRKKR-KPQAEKVDVIAGSSKMKGFSSSESESSSESSSSD 713
Query: 297 DDLNLFDALPISALL---------------KQSSNPLTPIPESQPAAQTTSPPHSPRSSF 341
+ + + P S +Q P+P+ P PP P
Sbjct: 714 SEDSETEMAPKSKKKGHPGREQKKHHHHHHQQMQQAPAPVPQQPPPPPQQPPPPPPPQQQ 773
Query: 342 FQPSPNEAP------LWNNFRTNPPD------PLIQPHPSPSKHQNVWLPELLIQTLPQ- 388
QP P P +++PP P+++P S + I LPQ
Sbjct: 774 QQPPPPPPPPSMPQQAAPAMKSSPPPFIATQVPVLEPQLPGSVFDPIGHFTQPILHLPQP 833
Query: 389 ---------PIHLYPSQL-MLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRY 438
P H P L + PP N L + + L P P R
Sbjct: 834 ELPPHLPQPPEHSTPPHLNQHAVVSPPALHNALPQQPSRPSNRAAALP------PKPARP 887
Query: 439 PGPRPERLVDP--DEPILANP----LHEADPLA-----QQVQPAPHQQQPVQPEPEPEQS 487
P P P +P +A P L + +P A Q+Q Q Q VQP S
Sbjct: 888 PAVSPALTQTPLLPQPPMAQPPQVLLEDEEPPAPPLTSMQMQLYLQQLQKVQPPTPLLPS 947
Query: 488 VSNHSSVRSPNP 499
V S P P
Sbjct: 948 VKVQSQPPPPLP 959
>PHC1_HUMAN (P78364) Polyhomeotic-like protein 1 (Early development
regulator protein 1) (HPH1)
Length = 1004
Score = 49.3 bits (116), Expect = 4e-05
Identities = 80/359 (22%), Positives = 139/359 (38%), Gaps = 48/359 (13%)
Query: 364 IQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLM------LLTLLPPITLNPLENSWRS 417
IQPH + Q + L + + + Q I ++ Q LL + L + +
Sbjct: 374 IQPHSLIQQQQQIHLQQKQV-VIQQQIAIHHQQQFQHRQSQLLHTATHLQLAQQQQQQQQ 432
Query: 418 EKEKVSTLEEYYLTCPSPRRYPGPR---PERLVDPDEPILANPLHEADPLAQQVQPAPHQ 474
++++ + LT P P + P + P + + ++ P+ ++ PL+ AP
Sbjct: 433 QQQQQQQPQATTLTAPQPPQVPPTQQVPPSQSQQQAQTLVVQPMLQSSPLSLPPDAAPKP 492
Query: 475 QQPVQPEP------EPEQSVSNHSSVRSPNPLVATSEPHLGASEP-HVQTCDIGSPQGAS 527
P+Q +P P+ + S+ + P P + +G +P Q +G Q A+
Sbjct: 493 PIPIQSKPPVAPIKPPQLGAAKMSAAQQPPPHIPVQV--VGTRQPGTAQAQALGLAQLAA 550
Query: 528 EAHSS----NHPASPETNLSIIPYTHLRPTSLSECINIFHQEASLMLCNVQGQTDLSENA 583
+S S + +L+ P + P +L EC + L VQG
Sbjct: 551 AVPTSRGMPGTVQSGQAHLASSPPSSQAPGALQECPPTL--APGMTLAPVQGTA------ 602
Query: 584 DYVDEEWHSLSTWLVAQVPV--IMQLLHAEGSQRIEAAKQRFARRVALHEQEQRQKLLEA 641
+V + + S+ +VAQVP MQ +H G + A K R+ E+ L +
Sbjct: 603 -HVVKGGATTSSPVVAQVPAAFYMQSVHLPGKPQTLAVK----RKADSEEERDDVSTLGS 657
Query: 642 IEEARRKQEQAEEAARLAAAQDEQARLEAERLEAEAEARRLAP----VVFTPVASASTP 696
+ A+ A A DE++ L E+ E+ A AP V TP S P
Sbjct: 658 MLPAK-----ASPVAESPKVMDEKSSL-GEKAESVANVNANAPSSELVALTPAPSVPPP 710
>YO62_DEIRA (Q9RRM6) Hypothetical UPF0144 protein DR2462
Length = 572
Score = 48.9 bits (115), Expect = 5e-05
Identities = 41/150 (27%), Positives = 69/150 (45%), Gaps = 17/150 (11%)
Query: 613 SQRIEAAKQRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQ----DEQ--- 665
+Q+I A QR AR + ++ RQ + ++A R+ + A E LAAA DEQ
Sbjct: 44 AQQIRAEAQRHARELHEAAEQDRQDAISKTQDAARRVQDAAERDTLAAAHEARLDEQREQ 103
Query: 666 -----ARLEAERLEAEAEARRLAPVVFTPVASASTPANQAAQNVP-----SSSTHSSSSR 715
A+LEAER +A+A+A + + T + + + + S +
Sbjct: 104 VRALRAQLEAEREQAKADAAQQREALSTDRQETRREREDLGREIERLGRRTEQLDARSDK 163
Query: 716 LDTVEQRLNTHESMLIEMKQMMMELLRRSD 745
LD +E+RL + +Q + E LR++D
Sbjct: 164 LDALEERLEGGWREVQRQEQEVAERLRQAD 193
>PCLO_RAT (Q9JKS6) Piccolo protein (Multidomain presynaptic
cytomatrix protein)
Length = 5085
Score = 47.8 bits (112), Expect = 1e-04
Identities = 84/393 (21%), Positives = 131/393 (32%), Gaps = 77/393 (19%)
Query: 203 ILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDNDDGPPSPK---KQKKQVRIV 259
++ DS + K K + + K + +KP PSPK KQ+ QV+
Sbjct: 174 LISDSEASQEETTKKQKVVQKEQGKSEGMAKPPLQQ--------PSPKPIPKQQGQVK-- 223
Query: 260 VKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDALPISALLKQSSNPLT 319
EVI++ +P V+ A+ K P + Q S T
Sbjct: 224 ----------EVIQQDSSPKSVSSQQAEKVKPQA------------PGTGKPSQQSPAQT 261
Query: 320 PIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLP 379
P ++ P P S +++ QP P ++P P QP P+ + Q L
Sbjct: 262 PAQQASPGKPVAQQPGSAKATVQQPGPAKSPAQPAGTGKSP---AQP-PAKTPGQQAGLE 317
Query: 380 ELLIQTLPQPIHL--------YP-SQLMLLTLLPPITLNPLENSWRSEKEKV----STLE 426
+ P P L +P + P +P + KV T
Sbjct: 318 KTSSSQQPGPKSLAQTPGHGKFPLGPVKSPAQQPGTAKHPAQQPGPQTAAKVPGPTKTPA 377
Query: 427 EYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQP------APHQQQPVQP 480
+ +P + PGP P +PI A P P+A + QP P QQP
Sbjct: 378 QQSGPGKTPAQQPGPTKP---SPQQPIPAKP-QPQQPVATKTQPQQSAPAKPQPQQPAPA 433
Query: 481 EPEPEQSVSNHSSVRSPNPLVATSEPHLG---------ASEPHVQTCDIGSPQGASEAHS 531
+P+P+Q + P P +P A++PH Q + P S
Sbjct: 434 KPQPQQPTPAKPQPQPPTPAKPQPQPPTATKPQPQPPTATKPHHQQPGLAKPSAQQPTKS 493
Query: 532 SNHPAS------PETNLSIIPYTHLRPTSLSEC 558
+ + P T+ + P L T C
Sbjct: 494 ISQTVTGRPLQPPPTSAAQTPAQGLSKTICPLC 526
Score = 35.0 bits (79), Expect = 0.72
Identities = 56/245 (22%), Positives = 95/245 (37%), Gaps = 34/245 (13%)
Query: 195 TKKVQRKMILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDG-----DDGDNDDGPPSP 249
T + R IL+D E D+ + K + + + DE K D + G N
Sbjct: 3700 TNTMARAKILQDIDRELDLVERESAKLRKKQAELDEEEKEIDAKLRYLEMGINRRKEALL 3759
Query: 250 KKQKKQVRIVVKPTRVEPAAEVIRRVET--PARV-TRSSAQSSKSAVASDDDLNLFDALP 306
K+++K+ R ++ + V + P+RV ++ + ++A ++ +
Sbjct: 3760 KEREKRERAYLQGVAEDRDYMSDSEVSSTRPSRVESQHGVERPRTAPQTEFSQFIPPQTQ 3819
Query: 307 ISALLKQSSNPLTPIPESQPAAQTTSP-PHSPRSSFFQP-------SPNEA-PLWNNFRT 357
A L ++P T S PA T +P P++ +S F Q SP + P + T
Sbjct: 3820 TEAQLVPPTSPYTQYQYSSPALPTQAPTPYTQQSHFQQQTLYHQQVSPYQTQPTFQAVAT 3879
Query: 358 NPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRS 417
P QP P+P QP + PSQ+M++ P T LE S
Sbjct: 3880 MSFTPQAQPTPTP-----------------QPSYQLPSQMMVIQQKPRQTTLYLEPKITS 3922
Query: 418 EKEKV 422
E +
Sbjct: 3923 NYEVI 3927
>P300_HUMAN (Q09472) E1A-associated protein p300 (EC 2.3.1.48)
Length = 2414
Score = 47.0 bits (110), Expect = 2e-04
Identities = 80/367 (21%), Positives = 144/367 (38%), Gaps = 58/367 (15%)
Query: 189 EFDPPLTKKVQRKMILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDNDDGP-P 247
E + ++Q++ +L +++ V +N G GQ + G +GP P
Sbjct: 643 ELEEKRRTRLQKQNMLPNAAGMVPVSMNPGPNMGQPQ-------------PGMTSNGPLP 689
Query: 248 SPKKQKKQVRIVVKPTRVEPAAEV-----IRRVETPARVTRSSAQSSKSAVASDDDLNLF 302
P + V + P R+ P + + + + P ++ +A LN
Sbjct: 690 DPSMIRGSVPNQMMP-RITPQSGLNQFGQMSMAQPPIVPRQTPPLQHHGQLAQPGALN-- 746
Query: 303 DALPISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDP 362
+ ++Q SN +P++Q +Q + + P + PS +AP+ ++ P
Sbjct: 747 PPMGYGPRMQQPSNQGQFLPQTQFPSQGMNVTNIPLA----PSSGQAPVSQAQMSSSSCP 802
Query: 363 LIQP-HPSPSKHQNVWLPEL----LIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRS 417
+ P P S+ ++ P+L L Q P P+ PS+ PP S +
Sbjct: 803 VNSPIMPPGSQGSHIHCPQLPQPALHQNSPSPV---PSRTPTPHHTPP--------SIGA 851
Query: 418 EKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPA-PHQQQ 476
++ +T+ T P+ PGP+ + L P P L QQVQP+ P
Sbjct: 852 QQPPATTIPAPVPTPPAMP--PGPQSQALHPPPRQ---TPTPPTTQLPQQVQPSLPAAPS 906
Query: 477 PVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPA 536
QP+ +P S +SV +PN + +P S+P V + E SN P+
Sbjct: 907 ADQPQQQPRSQQSTAASVPTPNAPLLPPQPATPLSQPAV----------SIEGQVSNPPS 956
Query: 537 SPETNLS 543
+ T ++
Sbjct: 957 TSSTEVN 963
Score = 35.8 bits (81), Expect = 0.42
Identities = 57/263 (21%), Positives = 88/263 (32%), Gaps = 37/263 (14%)
Query: 304 ALPISALLKQSSNPLTPIPES--QPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPD 361
A P + +Q + P TP P S QP + PP+ PR+ P PP
Sbjct: 1853 ATPTTPTGQQPTTPQTPQPTSQPQPTPPNSMPPYLPRTQAAGPVSQGKAAGQVTPPTPPQ 1912
Query: 362 ----PLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRS 417
PL P P P+ + + +T Q H+ Q + +PP+T
Sbjct: 1913 TAQPPL--PGPPPTAVEMAMQIQRAAETQRQMAHVQIFQRPIQHQMPPMT---------- 1960
Query: 418 EKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQP 477
+ + P R P E + P + + P +Q P P Q Q
Sbjct: 1961 ------PMAPMGMNPPPMTRGPSGHLEPGMGP------TGMQQQPPWSQGGLPQPQQLQS 2008
Query: 478 VQPEPEPEQSVSNHSSVRSPNPLVATSEPHLG-ASEPHVQTCDIGSPQGASEAHSSNHPA 536
P P SV+ H PL +P LG ++ + + + P+
Sbjct: 2009 GMPRP-AMMSVAQHG-----QPLNMAPQPGLGQVGISPLKPGTVSQQALQNLLRTLRSPS 2062
Query: 537 SPETNLSIIPYTHLRPTSLSECI 559
SP ++ H P L+ I
Sbjct: 2063 SPLQQQQVLSILHANPQLLAAFI 2085
Score = 33.1 bits (74), Expect = 2.7
Identities = 37/189 (19%), Positives = 64/189 (33%), Gaps = 33/189 (17%)
Query: 359 PPDPL--IQPHPSPSKHQNVW----LPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLE 412
PP P +Q H + N+ LP+ L + Y +L+ + P+ NP+
Sbjct: 2220 PPQPQQRMQHHMQQMQQGNMGQIGQLPQALGAEAGASLQAYQQRLLQQQMGSPVQPNPMS 2279
Query: 413 NSWRSEKEKVSTLEEYYLTCP--------SPRRYPGPRPERLVDPDEPILANPLHEADPL 464
+ + P SP+ P PRP+ + P H +
Sbjct: 2280 PQQHMLPNQAQSPHLQGQQIPNSLSNQVRSPQPVPSPRPQ----------SQPPHSSPSP 2329
Query: 465 AQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQ 524
Q QP+PH P P P + NP+ + H + + + + S
Sbjct: 2330 RMQPQPSPHHVSPQTSSPHPGLVAA------QANPM---EQGHFASPDQNSMLSQLASNP 2380
Query: 525 GASEAHSSN 533
G + H ++
Sbjct: 2381 GMANLHGAS 2389
>MAR3_LEIMA (P14700) Membrane antigen containing repeating peptides
(Clone 39) (Fragment)
Length = 53
Score = 47.0 bits (110), Expect = 2e-04
Identities = 28/53 (52%), Positives = 35/53 (65%), Gaps = 5/53 (9%)
Query: 631 EQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAE-----RLEAEAE 678
E E+ +L EEA R+Q +AEEAARL A +E ARL+AE RL+AEAE
Sbjct: 1 EAEEAARLQAEAEEAARQQAEAEEAARLQAEAEEAARLQAEAEEAARLQAEAE 53
>EXTN_TOBAC (P13983) Extensin precursor (Cell wall
hydroxyproline-rich glycoprotein)
Length = 620
Score = 46.6 bits (109), Expect = 2e-04
Identities = 62/270 (22%), Positives = 79/270 (28%), Gaps = 38/270 (14%)
Query: 247 PSPKKQKKQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDALP 306
PSP+ Q PT V+P R P T A + + LP
Sbjct: 194 PSPQVQPPPTYSPPPPTHVQPTPSPPSRGHQPQPPTHRHAPPTHRHAPPTHQPSPLRHLP 253
Query: 307 ISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQP 366
P P QP T SPP + QPSP +P + PP P+ P
Sbjct: 254 -------------PSPRRQPQPPTYSPPPPAYAQSPQPSPTYSPPPPTYSPPPPSPIYSP 300
Query: 367 HP---SPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVS 423
P SPS P T P Y PP T +P ++
Sbjct: 301 PPPAYSPSP------PPTPTPTFSPPPPAYS---------PPPTYSPPPPTY-------L 338
Query: 424 TLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPE 483
L + P P Y P P P L P + P P P +Q P P
Sbjct: 339 PLPSSPIYSPPPPVYSPPPPPSYSPPPPTYLPPPPPSSPPPPSFSPPPPTYEQSPPPPPA 398
Query: 484 PEQSVSNHSSVRSPNPLVATSEPHLGASEP 513
+ + P P + P P
Sbjct: 399 YSPPLPAPPTYSPPPPTYSPPPPTYAQPPP 428
Score = 44.3 bits (103), Expect = 0.001
Identities = 44/189 (23%), Positives = 64/189 (33%), Gaps = 7/189 (3%)
Query: 320 PIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLP 379
P P P T PP P S P P+ +P + +PP P P P+ + P
Sbjct: 357 PPPSYSPPPPTYLPPPPPSSP---PPPSFSPPPPTYEQSPPPPPAYSPPLPAPP--TYSP 411
Query: 380 ELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEE--YYLTCPSPRR 437
+ P P + P L PP +P S + Y P P
Sbjct: 412 PPPTYSPPPPTYAQPPPLPPTYSPPPPAYSPPPPPTYSPPPPTYSPPPPAYAQPPPPPPT 471
Query: 438 YPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSP 497
Y P P P PI + P + PL P P ++ + P P + + + P
Sbjct: 472 YSPPPPAYSPPPPSPIYSPPPPQVQPLPPTFSPPPPRRIHLPPPPHRQPRPPTPTYGQPP 531
Query: 498 NPLVATSEP 506
+P + P
Sbjct: 532 SPPTFSPPP 540
Score = 40.8 bits (94), Expect = 0.013
Identities = 48/193 (24%), Positives = 63/193 (31%), Gaps = 39/193 (20%)
Query: 320 PIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLP 379
P P P T SPP + P P +P + PP P+ P P + LP
Sbjct: 444 PPPTYSPPPPTYSPPPPAYAQPPPPPPTYSPPPPAYSPPPPSPIYSPPPPQVQP----LP 499
Query: 380 ELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYP 439
P+ IHL P PP R + T Y PSP +
Sbjct: 500 PTFSPPPPRRIHLPP---------PP---------HRQPRPPTPT----YGQPPSPPTFS 537
Query: 440 GPRPERLVDPDEPIL-------------ANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQ 486
P P ++ P P + P A P Q P P +QP P P Q
Sbjct: 538 PPPPRQIHSPPPPHWQPRTPTPTYGQPPSPPTFSAPPPRQIHSPPPPHRQPRPPTPTYGQ 597
Query: 487 SVSNHSSVRSPNP 499
S ++ P+P
Sbjct: 598 PPSPPTTYSPPSP 610
Score = 35.4 bits (80), Expect = 0.55
Identities = 48/199 (24%), Positives = 63/199 (31%), Gaps = 23/199 (11%)
Query: 317 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNV 376
P TPI P Q P + QP+P +PP QP P +H
Sbjct: 187 PPTPIYSPSPQVQPPPTYSPPPPTHVQPTP-----------SPPSRGHQPQPPTHRHAPP 235
Query: 377 WLPELLIQTLPQPI-HLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSP 435
P P+ HL PS P T +P ++ + T P P
Sbjct: 236 THRHAPPTHQPSPLRHLPPSP---RRQPQPPTYSPPPPAYAQSPQPSPTYS------PPP 286
Query: 436 RRYPGPRPERLVDPDEPILA-NPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSV 494
Y P P + P P + +P P PA + P P P S +
Sbjct: 287 PTYSPPPPSPIYSPPPPAYSPSPPPTPTPTFSPPPPA-YSPPPTYSPPPPTYLPLPSSPI 345
Query: 495 RSPNPLVATSEPHLGASEP 513
SP P V + P S P
Sbjct: 346 YSPPPPVYSPPPPPSYSPP 364
>NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 kDa
protein) (140 kDa nucleolar phosphoprotein) (Nopp140)
(Nucleolar and coiled-body phosphoprotein 1)
Length = 704
Score = 46.2 bits (108), Expect = 3e-04
Identities = 40/157 (25%), Positives = 66/157 (41%), Gaps = 20/157 (12%)
Query: 199 QRKMILEDSSEESDVP-----LNKGKKAGQSKRKHDETSKPDDGDDGDNDDGPPSPKKQK 253
Q+K + + + S VP L+K QS +K ++P D +++ S +++K
Sbjct: 288 QKKPMKKKAGPYSSVPPPSVSLSKKSVGAQSPKKAAAQTQPADSSADSSEESDSSSEEEK 347
Query: 254 KQVRIVVKPTRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDALPISALLKQ 313
K PA V+ + + A+SS + SD + A P+SA
Sbjct: 348 K-----------TPAKTVVSKTPAKPAPVKKKAESSSDSSDSDSSEDEAPAKPVSATKSP 396
Query: 314 SSNP-LTPIPESQPAAQTTSPPHSPRSSFFQPSPNEA 349
S P +TP P PAA+ + P P S +P +A
Sbjct: 397 LSKPAVTPKP---PAAKAVATPKQPAGSGQKPQSRKA 430
Score = 34.3 bits (77), Expect = 1.2
Identities = 46/190 (24%), Positives = 76/190 (39%), Gaps = 33/190 (17%)
Query: 175 IDVDEFFRMLPPAPEF----DPPLTKKVQRKMILEDSSEESD-------VPLNKG----- 218
+D+ F+ AP+ + P+ KK +++ DSSE+S VP K
Sbjct: 51 LDIYSFWLKSTKAPKVKLQSNGPVAKKAKKETSSSDSSEDSSEEEDKAQVPTQKAAAPAK 110
Query: 219 -----KKAGQSKRKHDETSKPDDGDDGDNDDGPPSPKKQKKQVRIVVKPTRVEPAAEVIR 273
+ AG++ K E+S ++ + + + K +KK+ V+ V+P A+ +R
Sbjct: 111 RASLPQHAGKAAAKASESSSSEESSEEEEE------KDKKKK---PVQQKAVKPQAKAVR 161
Query: 274 RVETPARVTRSSAQSSKSAVASDDDLNLFDALPISALLK-QSSNPLTPIP--ESQPAAQT 330
A + S + SS A A A K Q+ P P P ++QP A
Sbjct: 162 PPPKKAESSESESDSSSEDEAPQTQKPKAAATAAKAPTKAQTKAPAKPGPPAKAQPKAAN 221
Query: 331 TSPPHSPRSS 340
S SS
Sbjct: 222 GKAGSSSSSS 231
>MLL4_HUMAN (Q9UMN6) Myeloid/lymphoid or mixed-lineage leukemia
protein 4 (Trithorax homolog 2)
Length = 2715
Score = 46.2 bits (108), Expect = 3e-04
Identities = 62/250 (24%), Positives = 92/250 (36%), Gaps = 33/250 (13%)
Query: 305 LPISALLKQSSNPLTPIPESQPAA-----QTTSPPHSPRSSFF----QPSPNEAPLWNNF 355
+P + K+ PLTP ++ A + TSPP S+ + SP AP F
Sbjct: 461 VPATCSRKRGRPPLTPSQRAEREAARAGPEGTSPPTPTPSTATGGPPEDSPTVAPKSTTF 520
Query: 356 RTNPPDPLIQPHPSPSKHQNVWLPELLI-QTLPQPIHLYPSQLMLLTLLPPITLNPLENS 414
N I P S + + P + + P+P P + L PPIT +P
Sbjct: 521 LKNIRQ-FIMPVVSARSSRVIKTPRRFMDEDPPKP----PKVEVSPVLRPPITTSP---- 571
Query: 415 WRSEKEKVSTLEEYYLTCPSPRRYPGP--RPERLVDPDEPILANPLHEADPLAQQVQPAP 472
+ + PSP R P P P L + IL P L +++ P P
Sbjct: 572 ---------PVPQEPAPVPSPPRAPTPPSTPVPLPEKRRSILREPTFRWTSLTRELPPPP 622
Query: 473 HQQQPVQ-PEPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQG--ASEA 529
P P P P + S+ + P SE HL E + +G+P+
Sbjct: 623 PAPPPPPAPSPPPAPATSSRRPLLLRAPQFTPSEAHLKIYESVLTPPPLGAPEAPEPEPP 682
Query: 530 HSSNHPASPE 539
+ + PA PE
Sbjct: 683 PADDSPAEPE 692
Score = 40.8 bits (94), Expect = 0.013
Identities = 80/344 (23%), Positives = 115/344 (33%), Gaps = 54/344 (15%)
Query: 185 PPAPEFDPPLTKKVQRKMILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDNDD 244
PP P PP ++ Q + S VP +K G+ + ++ + G
Sbjct: 439 PPLPSPPPPPAQEEQ------EESPPPVVPATCSRKRGRPPLTPSQRAEREAARAGPEGT 492
Query: 245 GPPSPKKQKKQVRIVVKPTRVEPAAEV----IRRVETPARVTRSSAQSSKSAVASDDDLN 300
PP+P V P + IR+ P RSS D+D
Sbjct: 493 SPPTPTPSTATGGPPEDSPTVAPKSTTFLKNIRQFIMPVVSARSSRVIKTPRRFMDEDPP 552
Query: 301 LFDALPISALLKQSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPP 360
+ +S +L+ PI S P Q +P PSP AP PP
Sbjct: 553 KPPKVEVSPVLRP------PITTSPPVPQEPAPV---------PSPPRAP-------TPP 590
Query: 361 DPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKE 420
P P P K +++ L +P + S L LPP P S
Sbjct: 591 ST---PVPLPEKRRSI---------LREPTFRWTS---LTRELPPPPPAPPPPPAPSPPP 635
Query: 421 KVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQP 480
+T L +P+ P ++ E +L P A P A + +P P P +P
Sbjct: 636 APATSSRRPLLLRAPQFTPSEAHLKIY---ESVLTPPPLGA-PEAPEPEPPPADDSPAEP 691
Query: 481 EPEPEQSVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQ 524
EP +NH S+ P+V T P PH PQ
Sbjct: 692 EPR-AVGRTNHLSLPRFAPVVTT--PVKAEVSPHGAPALSNGPQ 732
>TOLA_ECOLI (P19934) TolA protein
Length = 421
Score = 45.8 bits (107), Expect = 4e-04
Identities = 35/114 (30%), Positives = 59/114 (51%), Gaps = 4/114 (3%)
Query: 610 AEGSQRIEAAKQRFARRVA---LHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQA 666
AE + +AA+Q +++ L QEQ+++ EA ++A KQ+QAEEAA AAA D +A
Sbjct: 89 AEELREKQAAEQERLKQLEKERLAAQEQKKQAEEAAKQAELKQKQAEEAAAKAAA-DAKA 147
Query: 667 RLEAERLEAEAEARRLAPVVFTPVASASTPANQAAQNVPSSSTHSSSSRLDTVE 720
+ EA+ AE A++ A + + A AQ ++ + + + E
Sbjct: 148 KAEADAKAAEEAAKKAAADAKKKAEAEAAKAAAEAQKKAEAAAAALKKKAEAAE 201
Score = 37.7 bits (86), Expect = 0.11
Identities = 34/130 (26%), Positives = 57/130 (43%), Gaps = 10/130 (7%)
Query: 614 QRIEAAKQRFARRVALHEQEQRQKLLE-----AIEEARRKQEQ----AEEAARLAAAQDE 664
+R+ A +Q+ A + E +QK E A +A+ K E AEEAA+ AAA D
Sbjct: 109 ERLAAQEQKKQAEEAAKQAELKQKQAEEAAAKAAADAKAKAEADAKAAEEAAKKAAA-DA 167
Query: 665 QARLEAERLEAEAEARRLAPVVFTPVASASTPANQAAQNVPSSSTHSSSSRLDTVEQRLN 724
+ + EAE +A AEA++ A + + A AA + ++ + ++
Sbjct: 168 KKKAEAEAAKAAAEAQKKAEAAAAALKKKAEAAEAAAAEARKKAATEAAEKAKAEAEKKA 227
Query: 725 THESMLIEMK 734
E + K
Sbjct: 228 AAEKAAADKK 237
>IF2_BRUSU (Q8FXT2) Translation initiation factor IF-2
Length = 959
Score = 45.8 bits (107), Expect = 4e-04
Identities = 33/65 (50%), Positives = 43/65 (65%), Gaps = 8/65 (12%)
Query: 624 ARRVALHEQEQRQ--KLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAE---RLEAEAE 678
ARR AL E + R+ + A+EEA+R+ AEE AR A ++E AR +AE RL+AEAE
Sbjct: 158 ARRRALEEAQIREVEERARAVEEAKRR---AEEDARRAKEREESARRQAEEEARLKAEAE 214
Query: 679 ARRLA 683
ARR A
Sbjct: 215 ARRKA 219
>MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2)
Length = 5179
Score = 45.4 bits (106), Expect = 5e-04
Identities = 57/242 (23%), Positives = 71/242 (28%), Gaps = 12/242 (4%)
Query: 316 NPLTPIPESQPAAQTTSPPHS----PRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPS 371
+P T P P TT PP + P ++ P P P T P P P P P
Sbjct: 1400 SPPTTTPSPPPTTTTTLPPTTTPSPPTTTTTTPPPTTTPSPPITTTTTPLPTTTPSP-PI 1458
Query: 372 KHQNVWLPELLIQ---TLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEY 428
P T P P PS T PP T P ST
Sbjct: 1459 STTTTPPPTTTPSPPTTTPSPPTTTPSPPTTTTTTPPPTTTPSPPMTTPITPPASTTTLP 1518
Query: 429 YLTCPSPRRYPGPRPERLVDPDEPILA--NPLHEADPLAQQVQPAPHQQQPVQPEPEPEQ 486
T PSP P P P P L P+P P P
Sbjct: 1519 PTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTTTP 1578
Query: 487 SVSNHSSVRSPNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPASPETNLSIIP 546
S ++ P T P S P T P + + + P +P T+ + +P
Sbjct: 1579 SPPTTTTPSPPTITTTTPPPTTTPSPPTTTTTT--PPPTTTPSPPTTTPITPPTSTTTLP 1636
Query: 547 YT 548
T
Sbjct: 1637 PT 1638
Score = 42.4 bits (98), Expect = 0.005
Identities = 54/238 (22%), Positives = 76/238 (31%), Gaps = 30/238 (12%)
Query: 319 TPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWL 378
TP P + P+ TT+ P P + P P P T P P P P +
Sbjct: 1571 TPPPTTTPSPPTTTTPSPPTITTTTPPPTTTPSPPTTTTTTPPPTTTPSPPTT---TPIT 1627
Query: 379 PELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRY 438
P TLP PS T PP T P + T PSP
Sbjct: 1628 PPTSTTTLPPT--TTPSPPPTTTTTPPPTTTPSPPT---------------TTTPSPPIT 1670
Query: 439 PGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPN 498
P P PI P + P P+ P P + + SS +P+
Sbjct: 1671 TTTTPPPTTTPSSPITTTP---SPPTTTMTTPS-----PTTTPSSPITTTTTPSSTTTPS 1722
Query: 499 PLVATSEPHLGASEPHVQTCDIGS-PQGASEAHSSNHPASPE-TNLSIIPYTHLRPTS 554
P T + P T + + P + + + P P T + P++ PT+
Sbjct: 1723 PPPTTMTTPSPTTTPSPPTTTMTTLPPTTTSSPLTTTPLPPSITPPTFSPFSTTTPTT 1780
Score = 42.4 bits (98), Expect = 0.005
Identities = 58/287 (20%), Positives = 87/287 (30%), Gaps = 30/287 (10%)
Query: 277 TPARVTRSSAQSSKSAVASDDDLNLFDALPISALLKQSSNPLTPIPESQPAAQTTSP--- 333
TP+ T ++ + S LP + S TP P + P+ TT+P
Sbjct: 1421 TPSPPTTTTTTPPPTTTPSPPITTTTTPLPTTTPSPPISTTTTPPPTTTPSPPTTTPSPP 1480
Query: 334 ---PHSPRSSFFQPSPNEAPLWNNFRTNPPDP-----LIQPHPSPSKHQNVWLPELLIQT 385
P P ++ P P P + T P P + P +PS P T
Sbjct: 1481 TTTPSPPTTTTTTPPPTTTP--SPPMTTPITPPASTTTLPPTTTPS-------PPTTTTT 1531
Query: 386 LPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRRYPGPRPER 445
P P PS + PP + L + +T T PSP P P
Sbjct: 1532 TPPPT-TTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPT 1590
Query: 446 LVDPDEPILANPLHEADPLA---QQVQPAPHQQQPVQPE------PEPEQSVSNHSSVRS 496
+ P P P+P P+ P P ++ +
Sbjct: 1591 ITTTTPPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTT 1650
Query: 497 PNPLVATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPASPETNLS 543
P P S P P + T P + + P+ P T ++
Sbjct: 1651 PPPTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITTTPSPPTTTMT 1697
>IF2_BRUME (Q8YEB3) Translation initiation factor IF-2
Length = 959
Score = 45.4 bits (106), Expect = 5e-04
Identities = 33/65 (50%), Positives = 42/65 (63%), Gaps = 8/65 (12%)
Query: 624 ARRVALHEQEQRQ--KLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAE---RLEAEAE 678
ARR AL E + R+ + A+EEA+R+ AEE AR A +E AR +AE RL+AEAE
Sbjct: 158 ARRRALEEAQIREVEERARAVEEAKRR---AEEDARRAKEHEESARRQAEEEARLKAEAE 214
Query: 679 ARRLA 683
ARR A
Sbjct: 215 ARRKA 219
>NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130
kDa protein) (140 kDa nucleolar phosphoprotein)
(Nopp140) (Nucleolar and coiled-body phosphoprotein 1)
Length = 699
Score = 45.1 bits (105), Expect = 7e-04
Identities = 106/511 (20%), Positives = 184/511 (35%), Gaps = 66/511 (12%)
Query: 205 EDSSEESDVPLNKGKK--AGQSKRKHDETSKPDDGDDGDNDDGPPSPKKQKKQVRIVVKP 262
+D ++ P+ KG K A +K + D D ++D PP K QK ++ V
Sbjct: 136 DDEEDQKKQPVQKGVKPQAKAAKAPPKKAKSSDSDSDSSSEDEPP--KNQKPKITPVTVK 193
Query: 263 TRVEPAAEVIRRVETPARVTRSSAQSSKSAVASDDDLNLFDALPISALLKQSSNPLTPIP 322
+ + + R A +S+ SS S+ +S DD S K ++ P +P
Sbjct: 194 AQTKAPPKPARAAPKIANGKAASSSSSSSSSSSSDD---------SEEEKAAATPKKTVP 244
Query: 323 ESQ-----PAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVW 377
+ Q P T+P SS S E + P P PS
Sbjct: 245 KKQVVAKAPVKAATTPTRKSSSSEDSSSDEEEEQKKPMKNKPGPYSYAPPPSAPP----- 299
Query: 378 LPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRR 437
P+ + T P + Q P+E+S S E S+ EE +
Sbjct: 300 -PKKSLGTQPPKKAVEKQQ-------------PVESSEDSSDESDSSSEE--------EK 337
Query: 438 YPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSP 497
P + +P A E+ + + + P +P + S SN +V +
Sbjct: 338 KPPTKAVVSKATTKPPPAKKAAESSSDSSDSDSSEDDEAPSKPAGTTKNS-SNKPAVTTK 396
Query: 498 NPLV----ATSEPHLGASEPHVQTCDIGSPQGASEAHSSNHPASPETNLSIIPYTHLRPT 553
+P V A +P G + + D S SE SS+ + E ++ T + T
Sbjct: 397 SPAVKPAAAPKQPVGGGQKLLTRKADSSS----SEEESSS--SEEEKTKKMVATTKPKAT 450
Query: 554 SLSECINIFHQEASLMLCNVQGQTDLSENAD--YVDEEWHSLSTWLVAQVPVIMQLLHAE 611
+ +++ ++A QG D S ++D +EE S V + P + A
Sbjct: 451 A-KAALSLPAKQAP------QGSRDSSSDSDSSSSEEEEEKTSKSAVKKKPQKV-AGGAA 502
Query: 612 GSQRIEAAKQRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAE 671
S+ A K + + + ++ E ++ + QA +A +A + +
Sbjct: 503 PSKPASAKKGKAESSNSSSSDDSSEEEEEKLKGKGSPRPQAPKANGTSALTAQNGKAAKN 562
Query: 672 RLEAEAEARRLAPVVFTPVASASTPANQAAQ 702
E E E ++ A VV + N+AA+
Sbjct: 563 SEEEEEEKKKAAVVVSKSGSLKKRKQNEAAK 593
Score = 33.5 bits (75), Expect = 2.1
Identities = 98/527 (18%), Positives = 180/527 (33%), Gaps = 86/527 (16%)
Query: 218 GKKAGQSKRKHDETSKPDDGDDGDNDDGPPSPKKQKKQVRIVVKPTRVEPAAEVIRRVET 277
G A ++K+K + D ++ + GPP+ K R+ + P +
Sbjct: 72 GPVAKKAKKKASSSDSEDSSEEEEEVQGPPAKKAAVPAKRVGLPPGKA------------ 119
Query: 278 PARVTRSSAQSSKSAVASDDDLNLFDALPISALLKQSSNPLTPIPESQPAAQTTSPPHSP 337
++S SS DDD P+ +K P ++ P +S S
Sbjct: 120 ---AAKASESSSSEESRDDDDEEDQKKQPVQKGVK----PQAKAAKAPPKKAKSSDSDSD 172
Query: 338 RSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSPSKHQNVWLPELLIQTLPQPIHLYPSQL 397
SS +P N+ P + P K Q + P+P P
Sbjct: 173 SSSEDEPPKNQKP--------------KITPVTVKAQT--------KAPPKPARAAPKIA 210
Query: 398 MLLTLLPPITLNPLENSWRSEKEKVS-----TLEEYYLTCPSPRRYPGPRPERLVDPDEP 452
+ + +S SE+EK + T+ + + +P + P R E
Sbjct: 211 NGKAASSSSSSSSSSSSDDSEEEKAAATPKKTVPKKQVVAKAPVK-AATTPTRKSSSSED 269
Query: 453 ILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQSVSNHSSVRSPNPLVATSEPHLGASE 512
++ E + +P P+ P P P++S+ + P V +P
Sbjct: 270 SSSDE-EEEQKKPMKNKPGPYSYAPPPSAPPPKKSL----GTQPPKKAVEKQQP------ 318
Query: 513 PHVQTCDIGSPQGASEAHSSNHPASPETNLSIIPYTHLRP------TSLSECINIFHQEA 566
V++ + S + S SS P T + T P S S+ + E
Sbjct: 319 --VESSEDSSDESDS---SSEEEKKPPTKAVVSKATTKPPPAKKAAESSSDSSDSDSSED 373
Query: 567 SLMLCNVQGQTDLSENADYVDEEWHSLSTWLVAQVPVIMQLLHAEGSQRI---EAAKQRF 623
G T S N V + ++ + PV G Q++ +A
Sbjct: 374 DEAPSKPAGTTKNSSNKPAVTTKSPAVKPAAAPKQPV-------GGGQKLLTRKADSSSS 426
Query: 624 ARRVALHEQEQRQKLLEAIE-EARRK------QEQAEEAARLAAAQDEQARLEAERLEAE 676
+ E+E+ +K++ + +A K +QA + +R +++ + + E E +
Sbjct: 427 EEESSSSEEEKTKKMVATTKPKATAKAALSLPAKQAPQGSRDSSSDSDSSSSEEEEEKTS 486
Query: 677 AEARRLAPVVFTPVASASTPANQAAQNVPSSSTHSSSSRLDTVEQRL 723
A + P A+ S PA+ SS++ SS + E++L
Sbjct: 487 KSAVKKKPQKVAGGAAPSKPASAKKGKAESSNSSSSDDSSEEEEEKL 533
>EXT2_ARATH (Q9M1G9) Extensin 2 precursor (AtExt2) (Cell wall
hydroxyproline-rich glycoprotein 1) (HRGP1)
Length = 743
Score = 45.1 bits (105), Expect = 7e-04
Identities = 51/204 (25%), Positives = 71/204 (34%), Gaps = 21/204 (10%)
Query: 315 SNPLTPIPESQPAAQTTSPP-----HSPRSSFFQPSPN---EAPLWNNFRTNPPDPLIQP 366
S+P P P SPP SP ++ PSP ++P ++PP P P
Sbjct: 503 SSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVHYKSPPPPYVYSSPPPPYYSP 562
Query: 367 HPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLE 426
SP H P + + P P + ++ + PP + + S KV
Sbjct: 563 --SPKVHYKSPPPPYVYNSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKV---- 616
Query: 427 EYYLTCPSPRRYPGPRPERLVDPDEPILAN----PLHEADPLAQQVQPAPHQQQPVQPEP 482
YY + P P Y P P P + P + P P PH V P P
Sbjct: 617 -YYKSPPPPYVYSSPPPP-YYSPSPKVYYKSPPPPYYSPSPKVYYKSP-PHPHVCVCPPP 673
Query: 483 EPEQSVSNHSSVRSPNPLVATSEP 506
P S S +SP P + P
Sbjct: 674 PPCYSPSPKVVYKSPPPPYVYNSP 697
Score = 38.1 bits (87), Expect = 0.085
Identities = 48/199 (24%), Positives = 66/199 (33%), Gaps = 28/199 (14%)
Query: 315 SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQPHPSP---S 371
S+P P P + SPP P P P +P + +PP P + P P S
Sbjct: 378 SSPPPPYYSPSPKVEYKSPP-PPYVYSSPPPPTYSPSPKVYYKSPPPPYVYSSPPPPYYS 436
Query: 372 KHQNVWL---PELLIQTLPQPIHLYPS-QLMLLTLLPPITLNPLENSWRSEKEKVSTLEE 427
V+ P + + P P + PS ++ + PP + + S KV
Sbjct: 437 PSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKV----- 491
Query: 428 YYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPEPEPEQS 487
YY + P P Y P P P + P P P P P S
Sbjct: 492 YYKSPPPPYVYSSPPP-------------PYYSPSPKVYYKSPPPPYVYSSPPPPYYSPS 538
Query: 488 VSNHSSVRSPNPLVATSEP 506
H +SP P S P
Sbjct: 539 PKVH--YKSPPPPYVYSSP 555
>YDC9_SCHPO (Q10172) Hypothetical protein C25G10.09c in chromosome I
Length = 1794
Score = 44.3 bits (103), Expect = 0.001
Identities = 97/415 (23%), Positives = 142/415 (33%), Gaps = 63/415 (15%)
Query: 185 PPAPE-FDPPLTKKVQRKMILEDSSEESDVPLNKGKKAGQSKRKHDETSKPDDGDDGDND 243
P AP+ +PP VQ+ +E ++ +V + S + D
Sbjct: 1184 PAAPQTLNPPSVSTVQQSKPIESNTHTPEVKATSESPSASSNLE----------DRAARI 1233
Query: 244 DGPPSPKKQKKQVRIVVKPTR--VEPAAEVIRRVETP-ARVTRSSAQSSKSAVASDDDLN 300
+ ++ + +KP + A V TP A T Q K A A +
Sbjct: 1234 KAEAQRRMNERLAALGIKPRQKGTPSPAPVNSATSTPVAAPTAQQIQPGKQASAVSSN-- 1291
Query: 301 LFDALPISALLKQSSNPLTPIPESQ--------PAAQTTSPPHSPRSSFFQPSPNEAPLW 352
+ A+ S P +P Q P A P + S P P +APL
Sbjct: 1292 ------VPAVSASISTPPAVVPTVQHPQPTKQIPTAAVKDPSTTSTSFNTAPIPQQAPLE 1345
Query: 353 NNFRTNPPDPLIQPH-PSPSKHQ-----NVWLPELLIQTLPQPIHLYPSQLMLLTLLPPI 406
N F +P ++P P+ K Q NV P + QP++ PS + P
Sbjct: 1346 NQFSKMSLEPPVRPAVPTSPKPQIPDSSNVHAPPPPV----QPMNAMPSH-NAVNARPSA 1400
Query: 407 TLNPLENSWRSEKEKVSTLEEYYLTCP---SPRRYPG-----------PRPERLVDPDEP 452
S VS++E+ T P S PG P P V P +P
Sbjct: 1401 PERRDSFGSVSSGSNVSSIEDETSTMPLKASQPTNPGAPSNHAPQVVPPAPMHAVAPVQP 1460
Query: 453 ILANPLHEAD-PLAQQVQPAPHQQQPVQ----PEPEPEQSVSNH--SSVRSPNPLVATSE 505
+ A P + PAP Q P P P SV+ SSV +P +++
Sbjct: 1461 KAPGMVTNAPAPSSAPAPPAPVSQLPPAVPNVPVPSMIPSVAQQPPSSV-APATAPSSTL 1519
Query: 506 PHLGASEPHVQTCDIGSPQGASEAHSSNHPASPETNLSIIPYTHLRPTSLSECIN 560
P +S HV + +PQ S A S+ PA PY P + IN
Sbjct: 1520 PPSQSSFAHVPSPAPPAPQHPSAAALSSAPADNSMPHRSSPYAPQEPVQKPQAIN 1574
>V70K_EPMV (P20129) 70 kDa protein
Length = 649
Score = 44.3 bits (103), Expect = 0.001
Identities = 47/180 (26%), Positives = 71/180 (39%), Gaps = 30/180 (16%)
Query: 312 KQSSNPLTPIPESQPAAQTTS-----PPHSPRSSFFQPSPNEAPLWNNFRTNPPDPLIQP 366
++S +P +P P + S PPHSP S+ + S + N+ N P P + P
Sbjct: 461 ERSCDPESPTPTLGHKTTSLSRLPLPPPHSPHSAQDRASALATDVSNSETKNCPSPTVPP 520
Query: 367 HPSPSKHQNVWLPELLIQTLPQPIHLYPSQLMLLTLL----------PPITLNPLENSWR 416
P H + LP T P+ + PS L L T L P + +P +S
Sbjct: 521 -PFLPNHLHPLLPGTDPPTTPRQLSPSPSSLSLRTFLDSAVISCDSSPVLPPSPSPSSHS 579
Query: 417 SEKEKVSTLEEYYLTCPSPRRYP-GPRPERLVDPDEPILANPLHEADPLAQQVQPAPHQQ 475
S + +C SP R+P P P +D +L + DP P PH++
Sbjct: 580 SSS---------FQSCTSPPRFPCYPPPPSALD----LLFSSTPGTDPFPPPDTPPPHKR 626
>SPA1_MOUSE (P46062) Signal-induced proliferation associated protein 1
(Sipa-1) (GTPase-activating protein Spa-1)
Length = 1037
Score = 44.3 bits (103), Expect = 0.001
Identities = 72/323 (22%), Positives = 130/323 (39%), Gaps = 57/323 (17%)
Query: 378 LPELLIQTLPQPIHLYPSQLMLLTLLPPITLNPLENSWRSEKEKVSTLEEYYLTCPSPRR 437
LP+L +T Q + P + +T+LPP S + + S E Y L+ P R
Sbjct: 731 LPKLGPETAAQMLRSAPK--VCVTVLPPD---------ESGRPRRSFSELYMLSLKEPSR 779
Query: 438 YPGPRPER-------LVDPDEPILANPLHEADPLAQQVQPAPHQQQPVQPE-PEPEQSVS 489
GP P + ++ P + +L L ++ P P + E S+S
Sbjct: 780 RGGPEPVQDETGKLVILPPTKQLLHFCLKDSSS-----PPGPGDLTEERTEFLRTHNSLS 834
Query: 490 NHSSVRSPNPLVATSEPHL---GASEPHVQTCDIGSP-----QGASEAHSSNHPASPETN 541
+ SS+ P++ + P L + P D +P G+ +H + PE
Sbjct: 835 SGSSLSDEAPVLPNTTPDLLLVTTANPSAPGTDRETPPSQDQSGSPSSHEDTSDSGPELR 894
Query: 542 LSIIPYTHLRPTSLSECINIFHQEASLMLCNVQGQTDLSENADYVDEEWHSLSTWLVAQV 601
SI+P R SL I+ EA ++ +++EW S+S + +
Sbjct: 895 ASILP----RTLSLRNSISKIMSEA---------------GSETLEDEWQSISE-IASTC 934
Query: 602 PVIMQLLHAEGSQRIEAAKQRFARRVALHEQ----EQRQKLLEAIEEARRKQEQAEEAAR 657
I++ L EG E+ + A + + ++ LE++ ++ Q E+A R
Sbjct: 935 NTILESLSREGQPISESGDPKEALKCDSEPEPGSLSEKVSHLESMLWKLQEDLQREKADR 994
Query: 658 LAAAQDEQA-RLEAERLEAEAEA 679
A ++ ++ R +RL AE+E+
Sbjct: 995 AALEEEVRSLRHNNQRLLAESES 1017
>PLE1_HUMAN (Q15149) Plectin 1 (PLTN) (PCN) (Hemidesmosomal protein 1)
(HD1)
Length = 4684
Score = 44.3 bits (103), Expect = 0.001
Identities = 39/136 (28%), Positives = 65/136 (47%), Gaps = 7/136 (5%)
Query: 605 MQLLHAEGSQRIEAAKQRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDE 664
+QL +R++A ++ A V EQE +Q L + R + +AE AAR AA + E
Sbjct: 2173 LQLAQEAAQKRLQAEEKAHAFAVQQKEQELQQTLQQEQSVLDRLRGEAE-AARRAAEEAE 2231
Query: 665 QARLEAERLEAEA-----EARRLAPVVFTPVASASTPANQAAQNVPSSSTHSSSSRLDTV 719
+AR++AER A++ EA RL A A A AA+ + + ++ R
Sbjct: 2232 EARVQAEREAAQSRRQVEEAERLKQSA-EEQAQARAQAQAAAEKLRKEAEQEAARRAQAE 2290
Query: 720 EQRLNTHESMLIEMKQ 735
+ L ++ EM++
Sbjct: 2291 QAALRQKQAADAEMEK 2306
Score = 41.2 bits (95), Expect = 0.010
Identities = 35/121 (28%), Positives = 57/121 (46%), Gaps = 9/121 (7%)
Query: 556 SECINIFHQEASLMLCNVQGQTDLSENADYVDEEWHSLSTWLVAQVPVIMQLLHAEGSQR 615
SE + QE L Q+ LSE + E + Q ++ L + +
Sbjct: 2610 SEEMQTVQQEQLLQETQALQQSFLSEKDSLLQRER------FIEQEKAKLEQLFQDEVAK 2663
Query: 616 IEAAKQRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAERLEA 675
+ ++ R+ EQE RQ+L+ ++EEARR+Q +AEE R Q+E +LE +R +
Sbjct: 2664 AQQLREEQQRQQQQMEQE-RQRLVASMEEARRRQHEAEEGVR--RKQEELQQLEQQRRQQ 2720
Query: 676 E 676
E
Sbjct: 2721 E 2721
Score = 39.7 bits (91), Expect = 0.029
Identities = 28/69 (40%), Positives = 39/69 (55%), Gaps = 6/69 (8%)
Query: 615 RIEAAKQRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAERLE 674
RIEA R + L +++ Q+ L+ EEA + ++ AEEAARL+ A E ARL E
Sbjct: 2383 RIEAEN----RALILRDKDNTQRFLQ--EEAEKMKQVAEEAARLSVAAQEAARLRQLAEE 2436
Query: 675 AEAEARRLA 683
A+ R LA
Sbjct: 2437 DLAQQRALA 2445
Score = 38.5 bits (88), Expect = 0.065
Identities = 41/131 (31%), Positives = 55/131 (41%), Gaps = 16/131 (12%)
Query: 624 ARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAE--------RLEA 675
ARR A +E R ++A EA + + Q EEA RL + +EQA+ A+ R EA
Sbjct: 2223 ARRAAEEAEEAR---VQAEREAAQSRRQVEEAERLKQSAEEQAQARAQAQAAAEKLRKEA 2279
Query: 676 EAEARRLAPVVFTPVASASTPANQAAQNVPSSSTHSSSSRLDTVEQRLNTHESMLIE--- 732
E EA R A A A A + + VEQ L T L E
Sbjct: 2280 EQEAARRAQA--EQAALRQKQAADAEMEKHKKFAEQTLRQKAQVEQELTTLRLQLEETDH 2337
Query: 733 MKQMMMELLRR 743
K ++ E L+R
Sbjct: 2338 QKNLLDEELQR 2348
Score = 36.6 bits (83), Expect = 0.25
Identities = 37/134 (27%), Positives = 62/134 (45%), Gaps = 19/134 (14%)
Query: 608 LHAEGSQRIEAAKQRFARRVALHEQEQR-------QKLLEAIEEARRKQEQAEEAARLAA 660
L ++ +EAA+QR ++A E+ +R QK L A EEA R+++ A E
Sbjct: 2096 LRSKEQAELEAARQR---QLAAEEERRRREAEERVQKSLAAEEEAARQRKAALEEVERLK 2152
Query: 661 AQDEQARLEAERLEAEAEARRLAPVVFTPVASASTPANQAAQNVPSSSTHSSSSRLDTVE 720
A+ E+AR ER E E+ AR+L A A + Q + + + ++
Sbjct: 2153 AKVEEARSLRERAEQES-ARQL--------QLAQEAAQKRLQAEEKAHAFAVQQKEQELQ 2203
Query: 721 QRLNTHESMLIEMK 734
Q L +S+L ++
Sbjct: 2204 QTLQQEQSVLDRLR 2217
Score = 36.6 bits (83), Expect = 0.25
Identities = 42/182 (23%), Positives = 76/182 (41%), Gaps = 22/182 (12%)
Query: 573 VQGQTDLS-ENADYVDEEWHSLSTWLVAQVPVIMQLLHAEGSQRIEAAKQRFARRVALHE 631
+Q Q +L+ E A + E+ ++ L + + L AE +++E + + A R+ L
Sbjct: 2469 LQQQKELAQEQARRLQEDKEQMAQQLAEETQGFQRTLEAERQRQLEMSAE--AERLKLRV 2526
Query: 632 QEQRQKLLEAIEEARRKQEQAEEAA----RLAAAQDEQA----RLEAERLEAEAEARRLA 683
E + A E+A+R ++QAEE R A E+ LE +R +++ +A RL
Sbjct: 2527 AEMSRAQARAEEDAQRFRKQAEEIGEKLHRTELATQEKVTLVQTLEIQRQQSDHDAERLR 2586
Query: 684 PVVFTPVASASTPANQAAQNVPSSSTHSSSSRLDTVEQRLNTHESMLIEMKQMMMELLRR 743
+ A + + S + TV+Q E +L E + + L
Sbjct: 2587 EAI------AELEREKEKLQQEAKLLQLKSEEMQTVQQ-----EQLLQETQALQQSFLSE 2635
Query: 744 SD 745
D
Sbjct: 2636 KD 2637
Score = 35.0 bits (79), Expect = 0.72
Identities = 33/129 (25%), Positives = 56/129 (42%), Gaps = 9/129 (6%)
Query: 600 QVPVIMQLLHAEGSQRIEAAKQRFARRVALHE---QEQRQKLLEAIEEARRKQEQAEEAA 656
QV V ++ +++ + FA + A E QE+ + + EEA R+ +Q EA
Sbjct: 1699 QVQVALETAQRSAEAELQSKRASFAEKTAQLERSLQEEHVAVAQLREEAERRAQQQAEAE 1758
Query: 657 RLAAAQDEQARLEAERLEAEAEARRL---APVVFTPVASASTPANQAAQNVPSSSTHSSS 713
R A ++ + +LE +L+A EA RL A V + A A + + +
Sbjct: 1759 R--AREEAERQLERWQLKAN-EALRLRLQAEEVLQQKSLAQAEAEKQKEEAEREARRRGK 1815
Query: 714 SRLDTVEQR 722
+ V QR
Sbjct: 1816 AEEQAVRQR 1824
Score = 34.3 bits (77), Expect = 1.2
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 8/73 (10%)
Query: 606 QLLHAEGSQRIEAAKQRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQ 665
QL QR+ A ++ R + EQ+++LLE EE R Q +A AAA ++
Sbjct: 1836 QLAEGTAQQRLAAEQELIRLRAETEQGEQQRQLLE--EELARLQREA------AAATQKR 1887
Query: 666 ARLEAERLEAEAE 678
LEAE + AE
Sbjct: 1888 QELEAELAKVRAE 1900
Score = 34.3 bits (77), Expect = 1.2
Identities = 22/62 (35%), Positives = 32/62 (51%)
Query: 620 KQRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQARLEAERLEAEAEA 679
K A R+ L +E Q+ A EA +++E+AE AR +EQA + E E E E
Sbjct: 1774 KANEALRLRLQAEEVLQQKSLAQAEAEKQKEEAEREARRRGKAEEQAVRQRELAEQELEK 1833
Query: 680 RR 681
+R
Sbjct: 1834 QR 1835
Score = 32.3 bits (72), Expect = 4.7
Identities = 30/89 (33%), Positives = 42/89 (46%), Gaps = 11/89 (12%)
Query: 606 QLLHAEGSQRIEAAKQRFARRVALHEQEQRQKLLEAIEEARRKQEQAEEAARLAAAQDEQ 665
+L + A + A + E+ ++ LEA EA R +E AEEAARL A +E
Sbjct: 1893 ELAKVRAEMEVLLASKAKAEEESRSTSEKSKQRLEA--EAGRFRELAEEAARLRALAEEA 1950
Query: 666 ARL------EAERLEAEAE---ARRLAPV 685
R +A R AEAE A +LA +
Sbjct: 1951 KRQRQLAEEDAARQRAEAERVLAEKLAAI 1979
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.130 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 91,274,671
Number of Sequences: 164201
Number of extensions: 4301374
Number of successful extensions: 25087
Number of sequences better than 10.0: 726
Number of HSP's better than 10.0 without gapping: 104
Number of HSP's successfully gapped in prelim test: 662
Number of HSP's that attempted gapping in prelim test: 20695
Number of HSP's gapped (non-prelim): 3151
length of query: 747
length of database: 59,974,054
effective HSP length: 118
effective length of query: 629
effective length of database: 40,598,336
effective search space: 25536353344
effective search space used: 25536353344
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0234.16


