

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0230.5
(406 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
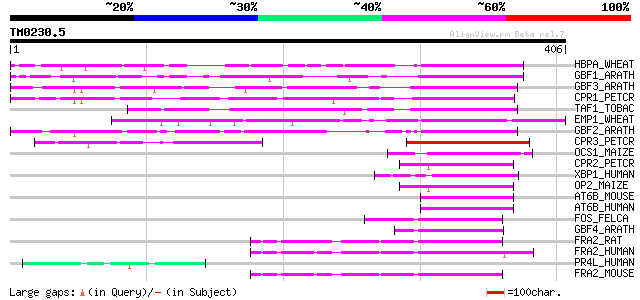
Score E
Sequences producing significant alignments: (bits) Value
HBPA_WHEAT (P23922) Transcription factor HBP-1a (Histone-specifi... 313 5e-85
GBF1_ARATH (P42774) G-box binding factor 1 (AtbZIP41) 181 3e-45
GBF3_ARATH (P42776) G-box binding factor 3 (AtbZIP55) 165 2e-40
CPR1_PETCR (Q99089) Common plant regulatory factor CPRF-1 164 3e-40
TAF1_TOBAC (Q99142) Transcriptional activator TAF-1 (Fragment) 136 1e-31
EMP1_WHEAT (P25032) DNA-binding EMBP-1 protein 129 1e-29
GBF2_ARATH (P42775) G-box binding factor 2 (AtbZIP54) 122 1e-27
CPR3_PETCR (Q99091) Light-inducible protein CPRF-3 88 4e-17
OCS1_MAIZE (P24068) Ocs-element binding factor 1 (OCSBF-1) 66 1e-10
CPR2_PETCR (Q99090) Light-inducible protein CPRF-2 57 1e-07
XBP1_HUMAN (P17861) X box binding protein-1 (XBP-1) (TREB5 protein) 54 9e-07
OP2_MAIZE (P12959) Opaque-2 regulatory protein 52 2e-06
AT6B_MOUSE (O35451) Cyclic-AMP-dependent transcription factor AT... 50 8e-06
AT6B_HUMAN (Q99941) Cyclic-AMP-dependent transcription factor AT... 50 1e-05
FOS_FELCA (Q8HZP6) Proto-oncogene protein c-fos (Cellular oncoge... 49 3e-05
GBF4_ARATH (P42777) G-box binding factor 4 (AtbZIP40) 48 4e-05
FRA2_RAT (P51145) Fos-related antigen 2 48 4e-05
FRA2_HUMAN (P15408) Fos-related antigen 2 47 7e-05
PR4L_HUMAN (P10162) Basic salivary proline-rich protein 4 allele... 47 9e-05
FRA2_MOUSE (P47930) Fos-related antigen 2 47 9e-05
>HBPA_WHEAT (P23922) Transcription factor HBP-1a (Histone-specific
transcription factor HBP1)
Length = 349
Score = 313 bits (802), Expect = 5e-85
Identities = 190/384 (49%), Positives = 229/384 (59%), Gaps = 68/384 (17%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNP--PDWSSFQAYSPMPPHGFL---A 55
MGS+ D + K SK P QEQPP T++GT P P+W FQ Y MPPHGF
Sbjct: 1 MGSN--DPSTPSKASKPPE---QEQPPATTSGTTAPVYPEWPGFQGYPAMPPHGFFPPPV 55
Query: 56 SNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIP---PGSYPFSPFAMPSPN 112
+ QAHPYMWG QH++PPYGTPP PY+ MYP G +YAHP+ P P YP P
Sbjct: 56 AAGQAHPYMWGPQHMVPPYGTPPPPYM-MYPPGTVYAHPTAPGVHPFHYPMQTNGNLEPA 114
Query: 113 GIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAH 172
G A G PG+ E +GK NE GKT G SANG
Sbjct: 115 G---AQGAAPGAAETNGK---------------------------NEPGKTSGPSANGVT 144
Query: 173 SKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTM 232
S S ESGS+ SEGSDANSQNDS K D E+ +QNG + H++ +G N P M
Sbjct: 145 SNS-ESGSDSESEGSDANSQNDSHSKEN---DVNENGSAQNGVS-HSSSHGTFNKP---M 196
Query: 233 SMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDG 292
+VP+ +G G V GPATNLNIGMDYWG SS +PA+ G VPS + G
Sbjct: 197 PLVPVQSGAVIG-VAGPATNLNIGMDYWGATGSSPVPAMRGKVPSGSARG---------- 245
Query: 293 VQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSR 352
+ W DERELK+Q+RK SNRESARRSRLRKQAEC+EL QRAEALK EN+SLR E+ R
Sbjct: 246 ---EQW--DERELKKQKRKLSNRESARRSRLRKQAECEELGQRAEALKSENSSLRIELDR 300
Query: 353 IRSDYEQLLTENAALKERLGELPG 376
I+ +YE+LL++N +LK +LGE G
Sbjct: 301 IKKEYEELLSKNTSLKAKLGESGG 324
>GBF1_ARATH (P42774) G-box binding factor 1 (AtbZIP41)
Length = 315
Score = 181 bits (459), Expect = 3e-45
Identities = 143/390 (36%), Positives = 186/390 (47%), Gaps = 110/390 (28%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDW-SSFQAY-----SPMPPHGFL 54
MG+SE DK P + + P + QE PPT PDW +S QAY +P P
Sbjct: 1 MGTSE-DKMPFK--TTKPTSSAQEVPPTPY------PDWQNSMQAYYGGGGTPNPFFPSP 51
Query: 55 ASNPQAHPYMWGVQH-IMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
+P HPYMWG QH +MPPYGTP PY AMYP G +YAHPS+P MP +G
Sbjct: 52 VGSPSPHPYMWGAQHHMMPPYGTPV-PYPAMYPPGAVYAHPSMP----------MPPNSG 100
Query: 114 IVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHG-KTPGTSANGAH 172
+ P +A GK +SKG+ ++K E G K S N
Sbjct: 101 ---PTNKEPAKDQASGK-----------KSKGN-------SKKKAEGGDKALSGSGNDGA 139
Query: 173 SKSGESGSEGTSEGSD--ANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQ 230
S S ES + G+S+ +D AN Q ++ D SQ+ T
Sbjct: 140 SHSDESVTAGSSDENDENANQQEQGSIRKPSFGQMLADASSQSTT--------------- 184
Query: 231 TMSMVPISAGGAPGAVP----GPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMAT 286
G G+VP P TNLNIGMD W + VP
Sbjct: 185 ---------GEIQGSVPMKPVAPGTNLNIGMDLWSS---------QAGVP---------- 216
Query: 287 GGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASL 346
++DERELKRQ+RKQSNRESARRSRLRKQAEC++L QR E+L EN SL
Sbjct: 217 ------------VKDERELKRQKRKQSNRESARRSRLRKQAECEQLQQRVESLSNENQSL 264
Query: 347 RSEVSRIRSDYEQLLTENAALKERLGELPG 376
R E+ R+ S+ ++L +EN ++++ L + G
Sbjct: 265 RDELQRLSSECDKLKSENNSIQDELQRVLG 294
>GBF3_ARATH (P42776) G-box binding factor 3 (AtbZIP55)
Length = 382
Score = 165 bits (418), Expect = 2e-40
Identities = 130/386 (33%), Positives = 181/386 (46%), Gaps = 75/386 (19%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYS----PMPPH---GF 53
MG+S + P K K PP T PDW++ QAY MPP+
Sbjct: 1 MGNSSEEPKPPTKSDKP------SSPPVDQTNVHVYPDWAAMQAYYGPRVAMPPYYNSAM 54
Query: 54 LASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGG-IYAHPSIPPGSYPFS----PFAM 108
AS PYMW QH+M PYG P Y A+YPHGG +YAHP IP GS P P
Sbjct: 55 AASGHPPPPYMWNPQHMMSPYGAP---YAAVYPHGGGVYAHPGIPMGSLPQGQKDPPLTT 111
Query: 109 PSPNGIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSA 168
P ++ + G+ + +G ++KE + S G+ G
Sbjct: 112 PGTLLSIDTPTKSTGNTD-NGLMKKLKEFDGLAMSLGN------------------GNPE 152
Query: 169 NGA--HSKSGESG-SEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGL 225
NGA H +S S ++G+++GSD N+ E K + S E P+++G + +
Sbjct: 153 NGADEHKRSRNSSETDGSTDGSDGNTTGADEPKL---KRSREGTPTKDGKQLVQASSFHS 209
Query: 226 NTPHQTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMA 285
+P + V + G PG + N N P S A+
Sbjct: 210 VSPSSGDTGVKLIQGSGAILSPGVSANSN--------PFMSQSLAM-------------- 247
Query: 286 TGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENAS 345
V + WLQ+ERELKR+RRKQSNRESARRSRLRKQAE +ELA++ EAL EN +
Sbjct: 248 -------VPPETWLQNERELKRERRKQSNRESARRSRLRKQAETEELARKVEALTAENMA 300
Query: 346 LRSEVSRIRSDYEQLLTENAALKERL 371
LRSE++++ ++L NA L ++L
Sbjct: 301 LRSELNQLNEKSDKLRGANATLLDKL 326
>CPR1_PETCR (Q99089) Common plant regulatory factor CPRF-1
Length = 411
Score = 164 bits (416), Expect = 3e-40
Identities = 122/383 (31%), Positives = 185/383 (47%), Gaps = 63/383 (16%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYS----PMPPH--GFL 54
MG+++ K K ++ +PP P P +++ PDW++ QAY +PP+ +
Sbjct: 1 MGNTDDVKAVKPEKLSSPP--PPAAPDQSNSHVY--PDWAAMQAYYGPRVALPPYFNPAV 56
Query: 55 ASNPQAHPYMWGV-QHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
AS HPYMWG Q +MPPYG P Y A+Y HGG+YAHP +P + P S
Sbjct: 57 ASGQSPHPYMWGPPQPVMPPYGVP---YAALYAHGGVYAHPGVPLAASPMS--------- 104
Query: 114 IVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHS 173
M+ K E IK+ KG I +GK + +
Sbjct: 105 -----------MDTHAKSSGTNEHGLIKKLKGHDDLAMSIG-----NGKADSSEGEMERT 148
Query: 174 KSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMS 233
S +EG+S+GS+ NS+ + GR+ ++ P+ G TQ+ + +P
Sbjct: 149 LSQSKETEGSSDGSNENSKRAAV---NGRKRGRDEAPNMIGEVKIETQSSVIPSPRAKSE 205
Query: 234 MV-------PISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMAT 286
+ P+ AG G V P+ ++ + + N PA G PST +
Sbjct: 206 KLLGITVATPMVAGKVVGTVVSPSMTSSLELKDSPKEHAVNSPA-GGQQPSTMMP----- 259
Query: 287 GGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASL 346
+ WL ++R+LKR+RRKQSNRESARRSRLRKQAE +ELA + ++L EN +L
Sbjct: 260 --------NDSWLHNDRDLKRERRKQSNRESARRSRLRKQAEAEELAIKVDSLTAENMAL 311
Query: 347 RSEVSRIRSDYEQLLTENAALKE 369
++E++R+ E+L +N+ L E
Sbjct: 312 KAEINRLTLTAEKLTNDNSRLLE 334
>TAF1_TOBAC (Q99142) Transcriptional activator TAF-1 (Fragment)
Length = 265
Score = 136 bits (342), Expect = 1e-31
Identities = 94/289 (32%), Positives = 140/289 (47%), Gaps = 33/289 (11%)
Query: 87 HGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGS 146
HGG+YAHP +P GS+P SP + +A S++A K E ++ + S G+
Sbjct: 2 HGGVYAHPGVPIGSHPPGHGMATSP-AVSQAMDGASLSLDASAKSSENSDRGLLAMSLGN 60
Query: 147 LGSLNMITRKNNEHGKTPGTSANGAHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSF 206
G H S +E +++GSD N SE R++
Sbjct: 61 --------------GSADNIEGGADHGNSQSGDTEDSTDGSDTNGAGVSERSKKRSRETT 106
Query: 207 EDEPSQNGTAVHTTQNGGLNTPHQTMSMVPISAGGAP----GAVPGPATNLNIGMDYWGT 262
D + + + Q G ++V + G G V P+ + M
Sbjct: 107 PDNSGDSKSHLRRCQPTGEINDDSEKAIVAVRPGKVGEKVMGTVLSPSMTTTLEMR---N 163
Query: 263 PTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSR 322
P S+++ A NV + A + ++ WLQ+ERELKR++RKQSNRESARRSR
Sbjct: 164 PASTHLKASPTNVSQLSPA-----------LPNEAWLQNERELKREKRKQSNRESARRSR 212
Query: 323 LRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALKERL 371
LRKQAE +ELA R ++L EN +L+SE++++ + E+L ENAAL ERL
Sbjct: 213 LRKQAEAEELAIRVQSLTAENMTLKSEINKLMENSEKLKLENAALMERL 261
>EMP1_WHEAT (P25032) DNA-binding EMBP-1 protein
Length = 354
Score = 129 bits (324), Expect = 1e-29
Identities = 112/349 (32%), Positives = 160/349 (45%), Gaps = 35/349 (10%)
Query: 75 GTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMP---SPNGIVEASGNTP---GSMEAD 128
GTP + Y + +P++ + +P +G+V A G++
Sbjct: 20 GTPAQAHAEWAASMHAYYAAAASAAGHPYAAWPLPPQAQQHGLVAAGAGAAYGAGAVPHV 79
Query: 129 GKPPEVKEKLPIKRSKG---SLGSLNMITRKNNEHGKT---PGTSANGAHSKSGESGSEG 182
PP + G G K GKT P N + S SG++GS+G
Sbjct: 80 PPPPAGTRHAHASMAAGVPYMAGESASAAGKGKRVGKTQRVPSGEINSS-SGSGDAGSQG 138
Query: 183 TSEGSDANSQNDSELKSGGRRDS----FEDEPSQNGTAVHTTQNGGLNTPHQTMSMVPIS 238
+SE DA + S RR S E EPSQ T + L ++ S + +
Sbjct: 139 SSEKGDAGANQKGSSSSAKRRKSGAAKTEGEPSQAATVQNAVTEPPLEDKERSASKLLVL 198
Query: 239 AGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHW 298
A G A+ A NLNIGMD P S++ PS+ V G + S S
Sbjct: 199 APGR-AALTSAAPNLNIGMD----PLSAS--------PSSLVQGEVNAAASSQSNASLSQ 245
Query: 299 LQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYE 358
+ DERELKR+RRKQSNRESARRSRLRKQ EC+ELAQ+ L N +LRSE+ +++ D +
Sbjct: 246 M-DERELKRERRKQSNRESARRSRLRKQQECEELAQKVSELTAANGTLRSELDQLKKDCK 304
Query: 359 QLLTENAALKERLGELPGNDD-LRSGKSDQHTHDDAQQSGQTEAVQGGH 406
+ TEN K+ +G++ +DD ++ + + Q E QGGH
Sbjct: 305 TMETEN---KKLMGKILSHDDKMQQSEGPSVVTTLSIQVEAPEPHQGGH 350
>GBF2_ARATH (P42775) G-box binding factor 2 (AtbZIP54)
Length = 360
Score = 122 bits (307), Expect = 1e-27
Identities = 112/378 (29%), Positives = 170/378 (44%), Gaps = 69/378 (18%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYS------PMPPHGFL 54
MGS+E + + P++ S V DW++ QAY P + L
Sbjct: 1 MGSNEEGNPTNNSDKPSQAAAPEQ-----SNVHVYHHDWAAMQAYYGPRVGIPQYYNSNL 55
Query: 55 ASNPQAHPYMWGVQH-IMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
A PYMW +M PYG P P+ P GG+YAHP + GS P P + S +G
Sbjct: 56 APGHAPPPYMWASPSPMMAPYGAPYPPFC---PPGGVYAHPGVQMGSQPQGPVSQ-SASG 111
Query: 114 IVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHS 173
+ TP +++A + +K+ K G I+ NN+ G +S+ H
Sbjct: 112 VT-----TPLTIDAPANSAGNSDHGFMKKLKEFDGLAMSIS--NNKVGSAEHSSSE--HR 162
Query: 174 KSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMS 233
S S ++G+S GSD N+ + + R+ + + G P TM
Sbjct: 163 SSQSSENDGSSNGSDGNTTGGEQSRRKRRQQRSPSTGERPSSQNSLPLRGENEKPDVTM- 221
Query: 234 MVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGV 293
GTP V TA++ + G +GV
Sbjct: 222 ---------------------------GTP-----------VMPTAMSFQNSAG--MNGV 241
Query: 294 QSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRI 353
Q W +E+E+KR++RKQSNRESARRSRLRKQAE ++L+ + +AL EN SLRS++ ++
Sbjct: 242 P-QPW--NEKEVKREKRKQSNRESARRSRLRKQAETEQLSVKVDALVAENMSLRSKLGQL 298
Query: 354 RSDYEQLLTENAALKERL 371
++ E+L EN A+ ++L
Sbjct: 299 NNESEKLRLENEAILDQL 316
>CPR3_PETCR (Q99091) Light-inducible protein CPRF-3
Length = 296
Score = 87.8 bits (216), Expect = 4e-17
Identities = 48/90 (53%), Positives = 60/90 (66%)
Query: 291 DGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEV 350
DG+ + DERELKRQRRKQSNRESARRSRLRKQA+ DEL +R + L +EN LR +
Sbjct: 183 DGMLPDQRVNDERELKRQRRKQSNRESARRSRLRKQAKSDELQERLDNLSKENRILRKNL 242
Query: 351 SRIRSDYEQLLTENAALKERLGELPGNDDL 380
RI ++ +EN ++KE L G D L
Sbjct: 243 QRISEACAEVTSENHSIKEELLRNYGPDGL 272
Score = 68.6 bits (166), Expect = 3e-11
Identities = 58/172 (33%), Positives = 76/172 (43%), Gaps = 26/172 (15%)
Query: 19 PLTPQEQPPTTSTGTVNPPDWSSFQAY-SPMPPHGFLAS---NPQAHPYMWGVQH-IMPP 73
P + E+ P T+T P SS QAY P F AS +P HPYMW QH + P
Sbjct: 15 PASSVEEAPITTTPF--PDLLSSMQAYYGGAAPAAFYASTVGSPSPHPYMWRNQHRFILP 72
Query: 74 YGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNTPGSMEADGKPPE 133
YG P Y A++ GGI+ HP +P PN P S E K +
Sbjct: 73 YGIPMQ-YPALFLPGGIFTHPIVPT-----------DPN-------LAPTSGEVGRKISD 113
Query: 134 VKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSKSGESGSEGTSE 185
K + K+S G GS + K E+ K +S N S S E+G +G+ E
Sbjct: 114 EKGRTSAKKSIGVSGSTSFAVDKGAENQKAASSSDNDCPSLSSENGVDGSLE 165
>OCS1_MAIZE (P24068) Ocs-element binding factor 1 (OCSBF-1)
Length = 151
Score = 66.2 bits (160), Expect = 1e-10
Identities = 41/106 (38%), Positives = 54/106 (50%), Gaps = 10/106 (9%)
Query: 277 STAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRA 336
S + G +G D H +R++R+ SNRESARRSRLRKQ DEL Q
Sbjct: 5 SLSPTAGRTSGSDGDSAADTH--------RREKRRLSNRESARRSRLRKQQHLDELVQEV 56
Query: 337 EALKEENASLRSEVSRIRSDYEQLLTENAALKERLGELPGNDDLRS 382
L+ +NA + + I S Y ++ EN L+ R EL D LRS
Sbjct: 57 ARLQADNARVAARARDIASQYTRVEQENTVLRARAAEL--GDRLRS 100
>CPR2_PETCR (Q99090) Light-inducible protein CPRF-2
Length = 401
Score = 56.6 bits (135), Expect = 1e-07
Identities = 37/92 (40%), Positives = 45/92 (48%), Gaps = 9/92 (9%)
Query: 286 TGGSRDGVQSQHWLQDEREL---------KRQRRKQSNRESARRSRLRKQAECDELAQRA 336
+G SRD L+ E E KR RR SNRESARRSR RKQA EL +
Sbjct: 171 SGSSRDHSDDDDELEGETETTRNGDPSDAKRVRRMLSNRESARRSRRRKQAHMTELETQV 230
Query: 337 EALKEENASLRSEVSRIRSDYEQLLTENAALK 368
L+ EN+SL ++ I Y +N LK
Sbjct: 231 SQLRVENSSLLKRLTDISQRYNDAAVDNRVLK 262
>XBP1_HUMAN (P17861) X box binding protein-1 (XBP-1) (TREB5 protein)
Length = 260
Score = 53.5 bits (127), Expect = 9e-07
Identities = 38/105 (36%), Positives = 54/105 (51%), Gaps = 6/105 (5%)
Query: 268 IPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQA 327
+PA G P A +GG+ R + H +E+ L RRK NR +A+ +R RK+A
Sbjct: 39 VPAQRGASPEAA-SGGLPQARKRQRLT--HLSPEEKAL---RRKLKNRVAAQTARDRKKA 92
Query: 328 ECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLG 372
EL Q+ L+EEN L E +R L+ EN L++RLG
Sbjct: 93 RMSELEQQVVDLEEENQKLLLENQLLREKTHGLVVENQELRQRLG 137
>OP2_MAIZE (P12959) Opaque-2 regulatory protein
Length = 453
Score = 52.4 bits (124), Expect = 2e-06
Identities = 32/91 (35%), Positives = 45/91 (49%), Gaps = 8/91 (8%)
Query: 286 TGGSRDGVQSQHWLQDEREL--------KRQRRKQSNRESARRSRLRKQAECDELAQRAE 337
+ SRD S + E E+ +R R+K+SNRESARRSR RK A EL +
Sbjct: 199 SSSSRDPSPSDEDMDGEVEILGFKMPTEERVRKKESNRESARRSRYRKAAHLKELEDQVA 258
Query: 338 ALKEENASLRSEVSRIRSDYEQLLTENAALK 368
LK EN+ L ++ + Y +N L+
Sbjct: 259 QLKAENSCLLRRIAALNQKYNDANVDNRVLR 289
>AT6B_MOUSE (O35451) Cyclic-AMP-dependent transcription factor ATF-6
beta (Activating transcription factor 6 beta)
(ATF6-beta) (cAMP responsive element binding
protein-like 1) (cAMP response element binding
protein-related protein) (Creb-rp)
Length = 699
Score = 50.4 bits (119), Expect = 8e-06
Identities = 29/68 (42%), Positives = 40/68 (58%)
Query: 301 DERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQL 360
D + LKRQ+R NRESA +SR +K+ L R +A+ +N LR E + +R E L
Sbjct: 319 DAKLLKRQQRMIKNRESACQSRRKKKEYLQGLEARLQAVLADNQQLRRENAALRRRLEAL 378
Query: 361 LTENAALK 368
L EN+ LK
Sbjct: 379 LAENSGLK 386
>AT6B_HUMAN (Q99941) Cyclic-AMP-dependent transcription factor ATF-6
beta (Activating transcription factor 6 beta)
(ATF6-beta) (cAMP responsive element binding
protein-like 1) (cAMP response element binding
protein-related protein) (Creb-rp) (G13 protein)
Length = 703
Score = 50.1 bits (118), Expect = 1e-05
Identities = 29/68 (42%), Positives = 40/68 (58%)
Query: 301 DERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQL 360
D + LKRQ+R NRESA +SR +K+ L R +A+ +N LR E + +R E L
Sbjct: 322 DAKLLKRQQRMIKNRESACQSRRKKKEYLQGLEARLQAVLADNQQLRRENAALRRRLEAL 381
Query: 361 LTENAALK 368
L EN+ LK
Sbjct: 382 LAENSELK 389
>FOS_FELCA (Q8HZP6) Proto-oncogene protein c-fos (Cellular oncogene
fos)
Length = 381
Score = 48.5 bits (114), Expect = 3e-05
Identities = 30/101 (29%), Positives = 51/101 (49%), Gaps = 2/101 (1%)
Query: 260 WGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESAR 319
+G P S V T AGG A R G Q L E E KR+ R++ N+ +A
Sbjct: 96 YGVPAPSAGAYSRAGVVKTVTAGGRAQSIGRRGKVEQ--LSPEEEEKRRIRRERNKMAAA 153
Query: 320 RSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQL 360
+ R R++ D L + L++E ++L++E++ + + E+L
Sbjct: 154 KCRNRRRELTDTLQAETDQLEDEKSALQTEIANLLKEKEKL 194
>GBF4_ARATH (P42777) G-box binding factor 4 (AtbZIP40)
Length = 270
Score = 48.1 bits (113), Expect = 4e-05
Identities = 33/81 (40%), Positives = 43/81 (52%), Gaps = 3/81 (3%)
Query: 282 GGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKE 341
GG T G R V + D+ +RQ+R NRESA RSR RKQA EL A L+E
Sbjct: 167 GGGVTRGKRGRVMME--AMDKAAAQRQKRMIKNRESAARSRERKQAYQVELETLAAKLEE 224
Query: 342 ENASLRSEVSR-IRSDYEQLL 361
EN L E+ + Y++L+
Sbjct: 225 ENEQLLKEIEESTKERYKKLM 245
>FRA2_RAT (P51145) Fos-related antigen 2
Length = 327
Score = 48.1 bits (113), Expect = 4e-05
Identities = 46/184 (25%), Positives = 82/184 (44%), Gaps = 12/184 (6%)
Query: 177 ESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMSMVP 236
++ S G+S GS A++++ S GG + F + +G+A T N T MV
Sbjct: 10 DTSSRGSS-GSPAHAESYSS--GGGGQQKFRVDMPGSGSAFIPTINAITTTSQDLQWMVQ 66
Query: 237 ISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQ 296
P + + Y P ++P H +P V + T R Q
Sbjct: 67 ------PTVITSMSNPYPRSHPYSPLPGLRSVPG-HMALPRPGVIKTIGTTVGRRRRDEQ 119
Query: 297 HWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSD 356
L E E KR+ R++ N+ +A + R R++ ++L E L+EE + L+ E++ ++ +
Sbjct: 120 --LSPEEEEKRRIRRERNKLAAAKCRNRRRELTEKLQTETEELEEEKSGLQKEIAELQKE 177
Query: 357 YEQL 360
E+L
Sbjct: 178 KEKL 181
>FRA2_HUMAN (P15408) Fos-related antigen 2
Length = 326
Score = 47.4 bits (111), Expect = 7e-05
Identities = 52/216 (24%), Positives = 95/216 (43%), Gaps = 22/216 (10%)
Query: 177 ESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMSMVP 236
++ S G+S GS A++++ S GG + F + +G+A T N + T MV
Sbjct: 10 DTSSRGSS-GSPAHAESYSS--GGGGQQKFRVDMPGSGSAFIPTINA-ITTSQDLQWMVQ 65
Query: 237 ISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQ 296
P + + Y P +++P H +P V + T R Q
Sbjct: 66 ------PTVITSMSNPYPRSHPYSPLPGLASVPG-HMALPRPGVIKTIGTTVGRRRRDEQ 118
Query: 297 HWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSD 356
L E E KR+ R++ N+ +A + R R++ ++L E L+EE + L+ E++ ++ +
Sbjct: 119 --LSPEEEEKRRIRRERNKLAAAKCRNRRRELTEKLQAETEELEEEKSGLQKEIAELQKE 176
Query: 357 YEQL---------LTENAALKERLGELPGNDDLRSG 383
E+L + + + + R PG +RSG
Sbjct: 177 KEKLEFMLVAHGPVCKISPEERRSPPAPGLQPMRSG 212
>PR4L_HUMAN (P10162) Basic salivary proline-rich protein 4 allele L
(Salivary proline-rich protein Po) (Parotid o protein)
[Contains: Peptide P-D] (Fragment)
Length = 276
Score = 47.0 bits (110), Expect = 9e-05
Identities = 42/137 (30%), Positives = 52/137 (37%), Gaps = 15/137 (10%)
Query: 10 PKEKESKTPPLTPQEQ-PPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQAHPYMWGVQ 68
P + ES+ P Q Q PP T PP Q+ PP G P+ P G Q
Sbjct: 83 PGKPESRPPQGGHQSQGPPPTPGKPEGPPPQGGNQSQGTPPPPG----KPEGRPPQGGNQ 138
Query: 69 HIMPPYGTPPHPYVAMYP--HGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNTPGSME 126
G PPHP P GG +H PP P P P G ++ G P +
Sbjct: 139 S----QGPPPHPGKPERPPPQGGNQSHRPPPPPGKP----ERPPPQGGNQSQGPPPHPGK 190
Query: 127 ADGKPPEVKEKLPIKRS 143
+G PP+ K RS
Sbjct: 191 PEGPPPQEGNKSRSARS 207
Score = 42.0 bits (97), Expect = 0.003
Identities = 51/192 (26%), Positives = 63/192 (32%), Gaps = 22/192 (11%)
Query: 18 PPLTPQEQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQAHPYMWGVQHIMPPYG-- 75
PP PQ PP + PP P P G + P HP G PP G
Sbjct: 19 PPGKPQGPPPQGGNQSQGPPPPPGKPEGRP-PQGGNQSQGPPPHP---GKPERPPPQGGN 74
Query: 76 ----TPPHPYV--AMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNTPGSMEADG 129
PPHP + P GG H S P P P P P G ++ G P + +G
Sbjct: 75 QSQGPPPHPGKPESRPPQGG---HQSQGPPPTPGKPEG-PPPQGGNQSQGTPPPPGKPEG 130
Query: 130 KPPEVKEKL------PIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSKSGESGSEGT 183
+PP+ + P K + N R GK G + G G
Sbjct: 131 RPPQGGNQSQGPPPHPGKPERPPPQGGNQSHRPPPPPGKPERPPPQGGNQSQGPPPHPGK 190
Query: 184 SEGSDANSQNDS 195
EG N S
Sbjct: 191 PEGPPPQEGNKS 202
>FRA2_MOUSE (P47930) Fos-related antigen 2
Length = 326
Score = 47.0 bits (110), Expect = 9e-05
Identities = 46/184 (25%), Positives = 84/184 (45%), Gaps = 13/184 (7%)
Query: 177 ESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMSMVP 236
++ S G+S GS A++++ S GG + F + +G+A T N + T MV
Sbjct: 10 DTSSRGSS-GSPAHAESYSS--GGGGQQKFRVDMPGSGSAFIPTINA-ITTSQDLQWMVQ 65
Query: 237 ISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQ 296
P + + Y P +++P H +P V + T R Q
Sbjct: 66 ------PTVITSMSNPYPRSHPYSPLPGLASVPG-HMALPRPGVIKTIGTTVGRRRRDEQ 118
Query: 297 HWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSD 356
L E E KR+ R++ N+ +A + R R++ ++L E L+EE + L+ E++ ++ +
Sbjct: 119 --LSPEEEEKRRIRRERNKLAAAKCRNRRRELTEKLQAETEELEEEKSGLQKEIAELQKE 176
Query: 357 YEQL 360
E+L
Sbjct: 177 KEKL 180
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.306 0.126 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 57,994,703
Number of Sequences: 164201
Number of extensions: 2943802
Number of successful extensions: 11651
Number of sequences better than 10.0: 819
Number of HSP's better than 10.0 without gapping: 204
Number of HSP's successfully gapped in prelim test: 629
Number of HSP's that attempted gapping in prelim test: 9542
Number of HSP's gapped (non-prelim): 2129
length of query: 406
length of database: 59,974,054
effective HSP length: 113
effective length of query: 293
effective length of database: 41,419,341
effective search space: 12135866913
effective search space used: 12135866913
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0230.5


