

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0228a.4
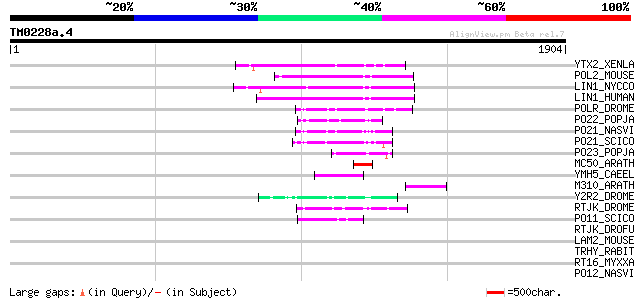
(1904 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 190 4e-47
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 189 8e-47
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 179 9e-44
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 174 2e-42
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 90 7e-17
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 73 7e-12
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 72 1e-11
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 70 7e-11
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 63 9e-09
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 63 9e-09
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 62 1e-08
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 62 1e-08
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 57 6e-07
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 55 2e-06
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 49 2e-04
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 45 0.003
LAM2_MOUSE (P21619) Lamin B2 36 1.2
TRHY_RABIT (P37709) Trichohyalin 35 1.5
RT16_MYXXA (P23072) RNA-directed DNA polymerase from retron Mx16... 35 2.0
PO12_NASVI (Q03270) Retrovirus-related Pol polyprotein from type... 35 2.0
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein (ORF
2)
Length = 1308
Score = 190 bits (482), Expect = 4e-47
Identities = 158/599 (26%), Positives = 276/599 (45%), Gaps = 30/599 (5%)
Query: 776 YSDHSPLLLRCH-APTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGNLQDKLQRVQE 834
+SDH+ + LR AP+ +AA + F + + F V +WR QD+ + +
Sbjct: 227 FSDHNCVSLRMSIAPSLPKAAY-WHFNNSLLEDEGFAKSVRDTWRGWRA-FQDEFATLNQ 284
Query: 835 -----------AATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKE 883
E+ K+V G + + + ++Q + ++L + ++
Sbjct: 285 WWDVGKVHLKLLCQEYTKSVSGQRNAEIEALNGEVLDLEQRLSGSEDQALQCEYLERKEA 344
Query: 884 YGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPST 943
++ ++ F +SR + DR ++FF+A + R +I LF EDG DP
Sbjct: 345 LRNMEQRQARGAFVRSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEA 404
Query: 944 LQEEAKRFYTNLFSLD-VQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSF 1002
+++ A+ FY NLFS D + + + P + +++L P++L+E+ A+ M
Sbjct: 405 IRDRARSFYQNLFSPDPISPDACEELWDGLPVVSERRKERLETPITLDELSQALRLMPHN 464
Query: 1003 KAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKD 1062
K+PG DG F++ +W LG D H ++ AF+ G +L L+PK + +K+
Sbjct: 465 KSPGLDGLTIEFFQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKN 524
Query: 1063 LRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTR 1122
RP+SL + YK++ K + R + +L +++ P Q + GR DN+ L ++++H R
Sbjct: 525 WRPVSLLSTDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLH--FAR 582
Query: 1123 KTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKL 1182
+TG L L +D EKA+DRV+ +L TL + F P+ V + + V N S
Sbjct: 583 RTGLSLAFLSLDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLT 642
Query: 1183 PSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADD 1242
RG+RQG PLS L+ L +E ++ + + + + + + +ADD
Sbjct: 643 APLAFGRGVRQGCPLSGQLYSLAIEPFLCLLRK-----RLTGLVLKEPDMRVVLSAYADD 697
Query: 1243 VLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQREL-SGLVGISR 1301
V+L Q D L + + AS ++N KS L G + L IS
Sbjct: 698 VILVAQDLVD-LERAQECQEVYAAASSARINWSKSSGLLE---GSLKVDFLPPAFRDISW 753
Query: 1302 ASNLGKYLGLPL-LQGRVKRDHFATIIEKVNTRLASWKN--NLLNKAGRVCLAKSILSS 1357
S + KYLG+ L + +F + E V TRL WK +L+ GR + +++S
Sbjct: 754 ESKIIKYLGVYLSAEEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRGRALVINQLVAS 812
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 189 bits (479), Expect = 8e-47
Identities = 137/486 (28%), Positives = 244/486 (50%), Gaps = 24/486 (4%)
Query: 909 RNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTNLFSLDVQV--RRNR 966
R TK + ++ + RN E G+ TDP +Q + FY L+S ++ ++
Sbjct: 405 RLTKGHRDKILINKIRN-------EKGDITTDPEEIQNTIRSFYKRLYSTKLENLDEMDK 457
Query: 967 IMEN-QTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGED 1025
++ Q P + ++ D L +P+S +E+ + I S+ + K+PGPDGF A FY+ + L
Sbjct: 458 FLDRYQVPKLNQDQVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPI 517
Query: 1026 LHHMVASAFQNGGANPELLATLLVLIPKVD-NPTKLKDLRPISLCNVSYKLITKVLVNRF 1084
LH + G + LIPK +PTK+++ RPISL N+ K++ K+L NR
Sbjct: 518 LHKLFHKIEVEGTLPNSFYEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRI 577
Query: 1085 RPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNW 1144
+ + ++ P Q FI G NI + V+H I+ K ++ + +D EKA+D++
Sbjct: 578 QEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYINKLKDKNHMI-ISLDAEKAFDKIQH 636
Query: 1145 DFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVL 1204
F+ L G +++I P ++ NG KL + PL+ G RQG PLSPYLF +
Sbjct: 637 PFMIKVLERSGIQGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNI 696
Query: 1205 CMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADF 1264
+E LA I+ ++ + + +++ K+ V IS L ADD++++ K+ R + + + F
Sbjct: 697 VLEVLARAIR---QQKEIKGIQIGKEEVKISLL--ADDMIVYISDPKNSTRELLNLINSF 751
Query: 1265 CEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQGRVK---RD 1321
E G K+N KS M F ++ ++E+ S +N KYLG+ L + VK
Sbjct: 752 GEVVGYKINSNKS-MAFLYTKNKQAEKEIRETTPFSIVTNNIKYLGVTLTK-EVKDLYDK 809
Query: 1322 HFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQS--LWLPDSVCNHVDKM 1379
+F ++ +++ L WK+ + GR+ + K + ++ + + +P N ++
Sbjct: 810 NFKSLKKEIKEDLRRWKDLPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGA 869
Query: 1380 VRNCIW 1385
+ +W
Sbjct: 870 ICKFVW 875
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 179 bits (453), Expect = 9e-44
Identities = 162/647 (25%), Positives = 299/647 (46%), Gaps = 43/647 (6%)
Query: 769 VEHLARVYSDHSPLLLRC------HAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQD 822
+E + ++SDH + + H T + WV + + T + Q+
Sbjct: 219 IEIIPCIFSDHHGIKVELNNNRNLHTHTKTWKLNNLMLKDTWVIDEIKKEI--TKFLEQN 276
Query: 823 GNLQDKLQRVQEAATEFNKTVFGNIHRRKRRVERR--------LQGIQQEHDWRDSESLA 874
N Q + + A + F + ++ ER L+ +++E S
Sbjct: 277 NNQDTNYQNLWDTAKAVLRGKFIALQAFLKKTEREEVNNLMGHLKQLEKEEHSNPKPSRR 336
Query: 875 RLERDLQKEYGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLED 934
+ ++ E +I + + KS+ +W F N + R+KR K + +
Sbjct: 337 KEITKIRAELNEIENKRIIQQINKSK-SWF-FEKINKIDKPLANLTRKKRVKSLISSIRN 394
Query: 935 GN--WCTDPSTLQEEAKRFYTNLFSLDVQVRR--NRIMEN-QTPTIPMEERDKLVAPVSL 989
GN TDPS +Q+ +Y L+S + + ++ +E P + +E + L P+S
Sbjct: 395 GNDEITTDPSEIQKILNEYYKKLYSHKYENLKEIDQYLEACHLPRLSQKEVEMLNRPISS 454
Query: 990 EEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATL-- 1047
E+ S I ++ K+PGPDGF + FY+ + E+L ++ + FQN L T
Sbjct: 455 SEIASTIQNLPKKKSPGPDGFTSEFYQTF----KEELVPILLNLFQNIEKEGILPNTFYE 510
Query: 1048 --LVLIPKVD-NPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRG 1104
+ LIPK +PT+ ++ RPISL N+ K++ K+L NR + + K++ Q FI G
Sbjct: 511 ANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQ 570
Query: 1105 TRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLI 1164
NI + V+ I+ K ++ L ID EKA+D + F+ TL G + LI
Sbjct: 571 GWFNIRKSINVIQHINKLKNKDHMI-LSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLI 629
Query: 1165 MWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRP 1224
P +++ NG KL SFPL+ G RQG PLSP LF + ME LAI I+ EE +
Sbjct: 630 EAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVMEVLAIAIR---EEKAIKG 686
Query: 1225 VRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKV 1284
+ + + + +S FADD++++ + ++D + + ++ SG K+N KS + F
Sbjct: 687 IHIGSEEIKLS--LFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKS-VAFIYT 743
Query: 1285 VGQRRQRELSGLVGISRASNLGKYLGLPLLQG--RVKRDHFATIIEKVNTRLASWKNNLL 1342
+ ++ + + + KYLG+ L + + ++++ T+ +++ + WKN
Sbjct: 744 NNNQAEKTVKDSIPFTVVPKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPC 803
Query: 1343 NKAGRVCLAKSILSSLPVHAMQSLWL--PDSVCNHVDKMVRNCIWGK 1387
+ GR+ + K + ++ ++ + P S ++K++ + IW +
Sbjct: 804 SWLGRINIVKMSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIWNQ 850
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 174 bits (442), Expect = 2e-42
Identities = 136/556 (24%), Positives = 275/556 (49%), Gaps = 21/556 (3%)
Query: 846 NIHRRKR---RVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEMLWFQKSREN 902
N H+RK+ +++ + +++ + S A +++ K ++ E QK E+
Sbjct: 302 NAHKRKQERSKIDTLISQLKELEKQEQTNSKASRRQEIIKIRAELKEIETQKTLQKINES 361
Query: 903 WVRFGDRNTKFFHAQTVV---RRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTNLFS-- 957
F ++ K + +R++N+I + + G+ TDP+ +Q + +Y +L++
Sbjct: 362 RSWFFEKINKIDRPLARLIKKKREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLYANK 421
Query: 958 LDVQVRRNRIMENQT-PTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYK 1016
L+ ++ ++ T P + EE + L P++ E+ + I S+ + K+PGP+GF A FY+
Sbjct: 422 LENLEEMDKFLDTYTLPRLNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQ 481
Query: 1017 QYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVD-NPTKLKDLRPISLCNVSYKL 1075
+Y L L + S + G ++LIPK + TK ++ RPISL N+ K+
Sbjct: 482 RYKEELVPFLLKLFQSIEKEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKI 541
Query: 1076 ITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDL 1135
+ K+L N+ + + KL+ Q FI NI + ++ I+ K ++ + ID
Sbjct: 542 LNKILANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKSINIIQHINRTKDTNHMI-ISIDA 600
Query: 1136 EKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGD 1195
EKA+D++ F+ L G + +I P +++ NG KL + PL+ G RQG
Sbjct: 601 EKAFDKIQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGC 660
Query: 1196 PLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLR 1255
PLSP L + +E LA I+ +E + + +++ K+ V +S FADD++++ + +
Sbjct: 661 PLSPLLPNIVLEVLARAIR---QEKEIKGIQLGKEEVKLS--LFADDMIVYLENPIVSAQ 715
Query: 1256 LIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQ 1315
+ +++F + SG K+N++KS+ F ++ + ++ + + AS KYLG+ L +
Sbjct: 716 NLLKLISNFSKVSGYKINVQKSQ-AFLYTNNRQTESQIMSELPFTIASKRIKYLGIQLTR 774
Query: 1316 G--RVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQS--LWLPDS 1371
+ ++++ ++ ++ WKN + GR+ + K + ++ + + LP +
Sbjct: 775 DVKDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRINIVKMAILPKVIYRFNAIPIKLPMT 834
Query: 1372 VCNHVDKMVRNCIWGK 1387
++K IW +
Sbjct: 835 FFTELEKTTLKFIWNQ 850
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 89.7 bits (221), Expect = 7e-17
Identities = 103/413 (24%), Positives = 177/413 (41%), Gaps = 37/413 (8%)
Query: 981 DKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGAN 1040
+++ + ++ +++R++ +S+ S +PGPDG K + + ++ G
Sbjct: 338 ERVWSAITEQDLRASRVSLSS--SPGPDGITP---KSAREVPSGIMLRIMNLILWCGNLP 392
Query: 1041 PELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFI 1100
+ V IPK + +D RPIS+ +V + + +L R ++ P Q F+
Sbjct: 393 HSIRLARTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSINW--DPRQRGFL 450
Query: 1101 LGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRT 1160
G DN + V+ H+ K +D+ KA+D ++ + TL +G P
Sbjct: 451 PTDGCADNATIVDLVLR--HSHKHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGF 508
Query: 1161 VDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEG 1220
VD + S+ +G F RG++QGDPLSP LF L M+RL + + +
Sbjct: 509 VDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEIG-- 566
Query: 1221 KWRPVRVSKDGVGISH-LFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRM 1279
+K G I++ FADD++LF + L+ TL DF G+K+N +K
Sbjct: 567 -------AKVGNAITNAAAFADDLVLFAETRMGLQVLLDKTL-DFLSIVGLKLNADKCFT 618
Query: 1280 LFSKVVGQRRQR-----ELSGLVGISRASNL-----GKYLGLPL-LQGRVKRDHFATIIE 1328
+ K GQ +Q+ S VG S +L KYLG+ GRV+ E
Sbjct: 619 VGIK--GQPKQKCTVLEAQSFYVGSSEIPSLKRTDEWKYLGINFTATGRVR----CNPAE 672
Query: 1329 KVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVCNHVDKMVR 1381
+ +L L R+ +++L H + + V DK++R
Sbjct: 673 DIGPKLQRLTKAPLKPQQRLFALRTVLIPQLYHKLALGSVAIGVLRKTDKLIR 725
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 73.2 bits (178), Expect = 7e-12
Identities = 73/293 (24%), Positives = 129/293 (43%), Gaps = 18/293 (6%)
Query: 986 PVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLA 1045
P++ EE++ AI K APG DG + + V G A
Sbjct: 5 PIAREEIQCAIKGWKP-SAPGSDGLTVQAITRT-----RLPRNFVQLHLLRGHVPTPWTA 58
Query: 1046 TLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGT 1105
LIPK + + RPI++ + +L+ ++L R + + P Q + GT
Sbjct: 59 MRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYARIDGT 116
Query: 1106 RDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIM 1165
N +L + + ++ +V+L D+ KA+D V+ + L G T + I
Sbjct: 117 LVNSLLLDTYISSRREQRKTYNVVSL--DVRKAFDTVSHSSICRALQRLGIDEGTSNYIT 174
Query: 1166 WGVRTPAVSV-LWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRP 1224
+ ++ + GS+ ++RG++QGDPLSP+LF ++ L +Q+ G
Sbjct: 175 GSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQSTPGIGG--- 231
Query: 1225 VRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKS 1277
+ ++ + + L FADD+LL + + L T+A+F GM +N +KS
Sbjct: 232 -TIGEEKIPV--LAFADDLLLL-EDNDVLLPTTLATVANFFRLRGMSLNAKKS 280
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 72.0 bits (175), Expect = 1e-11
Identities = 84/342 (24%), Positives = 145/342 (41%), Gaps = 28/342 (8%)
Query: 979 ERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGG 1038
E L P+S +E++ ++ A GPDG W+ + E + + +G
Sbjct: 311 EIKNLWRPISNDEIKEVEACKRT--AAGPDGMTT----TAWNSIDECIKSLFNMIMYHGQ 364
Query: 1039 ANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGS 1098
L + VLIPK RP+S+ +V+ + ++L NR H L+ Q +
Sbjct: 365 CPRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLDTRQRA 422
Query: 1099 FILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPP 1158
FI+ G +N L ++ R GL +D++KA+D V + L P
Sbjct: 423 FIVADGVAENTSLLSAMIK--EARMKIKGLYIAILDVKKAFDSVEHRSILDALRRKKLPL 480
Query: 1159 RTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVE 1218
+ IMW R + +K RG+RQGDPLSP LF M+ + ++ L E
Sbjct: 481 EMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCVMDAV---LRRLPE 537
Query: 1219 EGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSR 1278
+ + + +G L FADD++L + +++ L+ + + G+++ K
Sbjct: 538 NTGF---LMGAEKIGA--LVFADDLVLLAE-TREGLQASLSRIEAGLQEQGLEMMPRKCH 591
Query: 1279 MLFSKVVGQRRQRELSG----LVGISRASNLG-----KYLGL 1311
L G+ ++ ++ VG + LG KYLG+
Sbjct: 592 TLALVPSGKEKKIKVETHKPFTVGNQEITQLGHADQWKYLGV 633
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 69.7 bits (169), Expect = 7e-11
Identities = 92/357 (25%), Positives = 150/357 (41%), Gaps = 42/357 (11%)
Query: 970 NQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHM 1029
N+ P P E L PVSL E++SA S + K GPDG + W+ L + +
Sbjct: 144 NRAPPAPHMET--LWDPVSLIEIKSARASNE--KGAGPDGVTP----RSWNALDDRYKRL 195
Query: 1030 VASAFQNGGANPE-LLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLL 1088
+ + F G P + + V PK++ RP+S+C+V + K+L RF +
Sbjct: 196 LYNIFVFYGRVPSPIKGSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILARRF--VS 253
Query: 1089 HKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLR 1148
Q +++ G N+ + ++ ++ L +DL KA++ V L
Sbjct: 254 CYTYDERQTAYLPIDGVCINVSMLTAII--AEAKRLRKELHIAILDLVKAFNSVYHSALI 311
Query: 1149 ATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMER 1208
+ + G PP VD I + + G K + G+ QGDPLS LF L E+
Sbjct: 312 DAITEAGCPPGVVDYIADMYNNVITEMQFEG-KCELASILAGVYQGDPLSGPLFTLAYEK 370
Query: 1209 LAIQIQNLVEEGKW--RPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCE 1266
+ N EG++ VRV+ ++DD LL + ++H L F E
Sbjct: 371 ALRALNN---EGRFDIADVRVNASA-------YSDDGLLLAMT----VIGLQHNLDKFGE 416
Query: 1267 A---SGMKVNLEKSRMLF---------SKVVGQRRQRELSGLVGISRASNLGKYLGL 1311
G+++N KS+ + K+V RR + + S+L KYLG+
Sbjct: 417 TLAKIGLRINSRKSKTVSLVPSGREKKMKIVSNRRLLLKASELKPLTISDLWKYLGV 473
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 62.8 bits (151), Expect = 9e-09
Identities = 61/220 (27%), Positives = 92/220 (41%), Gaps = 26/220 (11%)
Query: 1104 GTRDNIILAQEVMHTIHTRKTGGGLV-ALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVD 1162
GT NII+ + H I R+ G + +D+ KA+D V+ + + FG D
Sbjct: 1 GTLANIIMLE---HYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQD 57
Query: 1163 LIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKW 1222
IM + +++ G ++ G++QGDPLSP LF + ++ L ++ +
Sbjct: 58 FIMSTITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASM 117
Query: 1223 RPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFS 1282
P I+ L FADD+LL D + T A F GM +N EK + +
Sbjct: 118 TP------ACKIASLAFADDLLLLEDRDIDVPNSLATTCAYF-RTRGMTLNPEKCASISA 170
Query: 1283 KVVGQRR-----------QRELSGLVGISRASNLGKYLGL 1311
V R R + L GI N KYLGL
Sbjct: 171 ATVSGRSVPRSKPSFTIDGRYIKPLGGI----NTFKYLGL 206
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 62.8 bits (151), Expect = 9e-09
Identities = 32/65 (49%), Positives = 41/65 (62%)
Query: 1178 NGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHL 1237
NG+ RGLRQGDPLSPYLF+LC E L+ + E+G+ +RVS + I+HL
Sbjct: 15 NGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRINHL 74
Query: 1238 FFADD 1242
FADD
Sbjct: 75 LFADD 79
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 62.4 bits (150), Expect = 1e-08
Identities = 46/169 (27%), Positives = 79/169 (46%), Gaps = 3/169 (1%)
Query: 1047 LLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTR 1106
+++ IPK NP+ + RPISL + +++ +++ +R R L+SP Q F+ R
Sbjct: 669 VIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCP 728
Query: 1107 DNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMW 1166
+++ + + H+I K L L D KA+D+V+ L L FG T
Sbjct: 729 SSLVRSISLYHSI--LKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKE 786
Query: 1167 GVRTPAVSVLWNG-SKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQ 1214
+ SV N ++P+ G+ QG P LF+L + L I ++
Sbjct: 787 FLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFILFINDLLIDLE 835
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 62.4 bits (150), Expect = 1e-08
Identities = 40/143 (27%), Positives = 64/143 (43%), Gaps = 2/143 (1%)
Query: 1357 SLPVHAMQSLWLPDSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAK-GRLGLRS 1415
+LPV+AM L +C + + W N R VAW+ + +SK G LG R
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 1416 ARKNNEALLGKLVHSFLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDS 1475
N+ALL K +++ L + L +Y S++ + SY WR I+ R+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 1476 VSDGFGPRLGAG-SSSLWYSNWL 1497
+S G +G G + +W W+
Sbjct: 122 LSRGLLRTIGDGIHTKVWLDRWI 144
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 56.6 bits (135), Expect = 6e-07
Identities = 110/495 (22%), Positives = 192/495 (38%), Gaps = 66/495 (13%)
Query: 852 RRVERRLQGIQQEHDWRDSESLARLE----RDLQKEYGDILYQEEMLWFQKSRENWVRF- 906
RR RRL+ + Q+ RD ++ L R Y ++ + +M ++W RF
Sbjct: 314 RREVRRLRRLLQDGRRRDDDAAVELVVVELRRASAYYKKLIGRAKM-------DDWKRFV 366
Query: 907 GDRNTKFFHAQTVVRRKRNKIH--GLFLEDGNWCTDPSTLQEEAKRFYTNLFSLDVQVRR 964
GD + + R R K G +G TD A+ N F
Sbjct: 367 GDHADDPWGRVYKICRGRRKCTEIGCLRVNGELITDWGDC---ARVLLRNFFP------- 416
Query: 965 NRIMENQTPTIPMEERDKLVAPVSLE--EVRSAIMSMKSFKAPGPDGFHAYFYKQYWHIL 1022
+ E++ PT EE P +LE EV + + +KS ++PG DG + K W +
Sbjct: 417 --VAESEAPTAIAEE-----VPPALEVFEVDTCVARLKSRRSPGLDGINGTICKAVWRAI 469
Query: 1023 GEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLK--DLRPISLCNVSYKLITKVL 1080
E L + + + G E +V + K + K + R I L V K++ ++
Sbjct: 470 PEHLASLFSRCIRLGYFPAEWKCPRVVSLLKGPDKDKCEPSSYRGICLLPVFGKVLEAIM 529
Query: 1081 VNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYD 1140
VNR R +L + Q F GR D A + + ++ +D + A+D
Sbjct: 530 VNRVREVLPE-GCRWQFGFRQGRCVED----AWRHVKSSVGASAAQYVLGTFVDFKGAFD 584
Query: 1141 RVNWDFLRATLYDFGFPPRTVDLIMW-GVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSP 1199
V W + L D G ++ +W + +V+ + S P+ RG QG P
Sbjct: 585 NVEWSAALSRLADLG----CREMGLWQSFFSGRRAVIRSSSGTVEVPVTRGCPQGSISGP 640
Query: 1200 YLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQA-SKDQLRLIK 1258
+++ + M+ L ++Q + +ADD+LL + S+ L
Sbjct: 641 FIWDILMDVLLQRLQPYCQLSA-----------------YADDLLLLVEGNSRAVLEEKG 683
Query: 1259 HTLADFCEASGMKVN--LEKSRMLFSKVVGQRRQRELSGLVGISRA-SNLGKYLGLPLLQ 1315
L E G +V L S+ + + G R+ G + +YLG+ + +
Sbjct: 684 AQLMSIVETWGAEVGDCLSTSKTVIMLLKGALRRAPTVRFAGRNLPYVRSCRYLGITVSE 743
Query: 1316 GRVKRDHFATIIEKV 1330
G H A++ +++
Sbjct: 744 GMKFLTHIASLRQRM 758
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 54.7 bits (130), Expect = 2e-06
Identities = 94/402 (23%), Positives = 167/402 (41%), Gaps = 44/402 (10%)
Query: 984 VAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQ---NGGAN 1040
++ V+LEEV++ I + KAPG D + +L + +A F + G
Sbjct: 435 LSAVTLEEVKNLIAKLPLKKAPGED----LLDNRTIRLLPDQALQFLALIFNSVLDVGYF 490
Query: 1041 PEL--LATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPL--LHKLVSPLQ 1096
P+ A+++++ PT + RP SL K++ ++++NR + K + Q
Sbjct: 491 PKAWKSASIIMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQ 550
Query: 1097 GSFILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFG- 1155
F L GT + L + V + + V +D+++A+DRV W LY
Sbjct: 551 FGFRLQHGTPEQ--LHRVVNFALEAMENKEYAVGAFLDIQQAFDRV-WH--PGLLYKAKR 605
Query: 1156 -FPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQ 1214
FPP+ ++ + V +G K P+ G+ QG L P L+ + +
Sbjct: 606 LFPPQLYLVVKSFLEERTFHVSVDGYKSSIKPIAAGVPQGSVLGPTLYSVFASDMPTH-- 663
Query: 1215 NLVEEGKWRPV-RVSKDGVGISHLFFADDVLLFCQ-----ASKDQLRLIKHTLADFCEAS 1268
PV V ++ V I+ +ADD + + A+ L+ + E
Sbjct: 664 --------TPVTEVDEEDVLIA--TYADDTAVLTKSKSILAATSGLQEYLDAFQQWAENW 713
Query: 1269 GMKVNLEK-SRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQGRVKRDHFATII 1327
+++N EK + + F+ G L+G + R KYLG+ L + H I
Sbjct: 714 NVRINAEKCANVTFANRTGSCPGVSLNG--RLIRHHQAYKYLGITLDRKLTFSRHITNIQ 771
Query: 1328 EKVNTRLA--SW---KNNLLNKAGRVCLAKSILSSLPVHAMQ 1364
+ T++A SW N L+ +V + KSIL+ + +Q
Sbjct: 772 QAFRTKVARMSWLIAPRNKLSLGCKVNIYKSILAPCLFYGLQ 813
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 48.5 bits (114), Expect = 2e-04
Identities = 50/231 (21%), Positives = 99/231 (42%), Gaps = 12/231 (5%)
Query: 987 VSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASA-FQNGGANPELLA 1045
+ ++EV ++ K K+PGPDG + W + E + + ++ +A
Sbjct: 405 LEMDEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIA 464
Query: 1046 TLLVLIPKVDN-PTKLKDLRPISLCNVSYKLITKVLVNRF-RPLLHKLVSPLQGSFILGR 1103
+L++L+ +D + RPI L + K++ ++V R + L+ VSP Q +F G+
Sbjct: 465 SLVILLKLLDRIRSDPGSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGK 524
Query: 1104 GTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDL 1163
T D Q + + ++ L ID + A+D + W + L + ++ +
Sbjct: 525 STEDAWRCVQRHVECSEMKY----VIGLNIDFQGAFDNLGWLSMLLKLDE----AQSNEF 576
Query: 1164 IMWGVRTPAVSVLWNG-SKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQI 1213
+W V + G + + + RG QG P ++ L M L + +
Sbjct: 577 GLWMSYFGGRKVYYVGKTGIVRKDVTRGCPQGSKSGPAMWKLVMNELLLAL 627
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 44.7 bits (104), Expect = 0.003
Identities = 131/645 (20%), Positives = 250/645 (38%), Gaps = 84/645 (13%)
Query: 769 VEHLARVYSDHSPLLLRCHAPTGDRAARPF---------RFQAAWVSH------------ 807
V+ L + SDH+PLL HA ++ R RF+A H
Sbjct: 204 VQTLHELSSDHTPLLADLHAMPINKPPRSCLLARGADIERFKAYLTQHIDLSVGIQGTDD 263
Query: 808 --PSFESVVNTSWRAQDGNLQDKLQRVQEAATEFNKTVFGNIHRRKRRVERRLQGIQQEH 865
+ +S ++ RA + Q V+ + + ++ R KR+V R +
Sbjct: 264 IDNAIDSFMDILKRAAIRSAPSHQQNVESSRQLQLPPIVASLIRLKRKVRREYA---RTG 320
Query: 866 DWRDSESLARLERDLQKEYGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRN 925
D R + +RL L K +L + + EN G N + + +
Sbjct: 321 DARIQQIHSRLANRLHK----VLNRRKQSQIDNLLENLDTDGSTNFSLWRITKRYKTQAT 376
Query: 926 KIHGLFLEDGNWCTDPSTLQEEAKRFYTNLF----SLDVQVRRNRIMENQTPTIPMEERD 981
+ G WC T +E+ + F +L +L + +M ++ P +
Sbjct: 377 PNSAIRNPAGGWCR---TSREKTEVFANHLEQRFKALAFAPESHSLMVAESLQTPFQMAL 433
Query: 982 KLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVA--SAFQNGGA 1039
PV+LEEV+ + +K KAPG D + + + L ++V ++ G
Sbjct: 434 P-ADPVTLEEVKELVSKLKPKKAPGEDLLDNRTIRL---LPDQALLYLVLIFNSILRVGY 489
Query: 1040 NPEL--LATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRF--RPLLHKLVSPL 1095
P+ A++++++ P + RP SL K++ ++++NR + + +
Sbjct: 490 FPKARPTASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRILTSEEVTRAIPKF 549
Query: 1096 QGSFILGRGTRDNI-ILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDF 1154
Q F L GT + + + + + ++ G +D+++A+DRV W LY
Sbjct: 550 QFGFRLQHGTPEQLHRVVNFALEALEKKEYAGSCF---LDIQQAFDRV-WH--PGLLYKA 603
Query: 1155 G--FPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQ 1212
P+ LI SV +G + ++ G+ QG L P L+ + +
Sbjct: 604 KSLLSPQLFQLIKSFWEGRKFSVTADGCRSSVKFIEAGVPQGSVLGPTLYSIFTADM--P 661
Query: 1213 IQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQA-----SKDQLRLIKHTLADFCEA 1267
QN V V ++ +ADD+ + ++ + D L+ ++
Sbjct: 662 NQNAVTGLAEGEVLIAT---------YADDIAVLTKSTCIVEATDALQEYLDAFQEWAVK 712
Query: 1268 SGMKVNLEK-SRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQGRVKRDH---- 1322
+ +N K + + F+ + ++G + ++ KYLG+ L + R H
Sbjct: 713 WNVSINAGKCANVTFTNAIRDCPGVTING--SLLSHTHEYKYLGVILDRSLTFRRHITSL 770
Query: 1323 ---FATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQ 1364
F T I K+N LA+ N L+ +V + K I++ +A+Q
Sbjct: 771 QHSFRTRITKMNWLLAA--RNKLSLDNKVKIYKCIVAPGLFYAIQ 813
>LAM2_MOUSE (P21619) Lamin B2
Length = 592
Score = 35.8 bits (81), Expect = 1.2
Identities = 31/101 (30%), Positives = 56/101 (54%), Gaps = 13/101 (12%)
Query: 824 NLQDKLQRVQEAATEFNKTVFGN-IHRRKRRVERRLQGI----QQEHDWRDSESLARLER 878
+L+++ Q +QE F+K+VF + +RR ERRL + QQE+D++ +++L
Sbjct: 185 DLENRCQSLQEELA-FSKSVFEEEVRETRRRHERRLVEVDSSRQQEYDFKMAQAL----E 239
Query: 879 DLQKEYGD--ILYQEEM-LWFQKSRENWVRFGDRNTKFFHA 916
DL+ ++ + LY+ E+ +Q +N D+N K HA
Sbjct: 240 DLRSQHDEQVRLYRVELEQTYQAKLDNAKLLSDQNDKAAHA 280
>TRHY_RABIT (P37709) Trichohyalin
Length = 1407
Score = 35.4 bits (80), Expect = 1.5
Identities = 30/132 (22%), Positives = 57/132 (42%), Gaps = 23/132 (17%)
Query: 851 KRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEMLWFQKSRENWVRFGDRN 910
+ R E RL+ ++ R+ E L R E ++ D ++EE Q+ E +R +R+
Sbjct: 966 QEREEERLRRQERARKLREEEQLLRREEQELRQERDRKFREEEQLLQEREEERLRRQERD 1025
Query: 911 TKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTNLFSLDVQVRRNRIMEN 970
KF + +RR+ L+E+ ++ F L+ Q+R+ E
Sbjct: 1026 RKFREEERQLRRQE-------------------LEEQFRQERDRKFRLEEQIRQ----EK 1062
Query: 971 QTPTIPMEERDK 982
+ + +ERD+
Sbjct: 1063 EEKQLRRQERDR 1074
Score = 33.9 bits (76), Expect = 4.4
Identities = 23/109 (21%), Positives = 52/109 (47%), Gaps = 2/109 (1%)
Query: 821 QDGNLQDKLQRVQEAATEFNKTVFGNIHRRKRRVERRLQGIQQEHD--WRDSESLARLER 878
++ L+++ Q +++ E + + ++ + + Q ++QE D R+ E L R E
Sbjct: 842 RERKLREEEQLLRQEEQELRQERARKLREEEQLLRQEEQELRQERDRKLREEEQLLRQEE 901
Query: 879 DLQKEYGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKI 927
++ D +EE Q+S E +R +R K + ++RR+ ++
Sbjct: 902 QELRQERDRKLREEEQLLQESEEERLRRQERERKLREEEQLLRREEQEL 950
>RT16_MYXXA (P23072) RNA-directed DNA polymerase from retron Mx162-RT
(EC 2.7.7.49) (Reverse transcriptase) (Mx162-RT)
Length = 485
Score = 35.0 bits (79), Expect = 2.0
Identities = 46/217 (21%), Positives = 85/217 (38%), Gaps = 42/217 (19%)
Query: 1094 PLQGS---FILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNWDFLRAT 1150
P+ G+ F+ GR N + Q G V +K+DL+ + V W ++
Sbjct: 219 PVHGAAHGFVAGRSILTNALAHQ------------GADVVVKVDLKDFFPSVTWRRVKGL 266
Query: 1151 LYDFGFPPRTVDLI-MWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERL 1209
L G T L+ + P +V + G L R L QG P SP + +L
Sbjct: 267 LRKGGLREGTSTLLSLLSTEAPREAVQFRGKLLHVAKGPRALPQGAPTSPGITNALCLKL 326
Query: 1210 AIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKD---------QLRLIKHT 1260
++ L + +G ++ +ADD+ +K + ++
Sbjct: 327 DKRLSALAKR------------LGFTYTRYADDLTFSWTKAKQPKPRRTQRPPVAVLLSR 374
Query: 1261 LADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLV 1297
+ + EA G +V+ +K+R V + ++ ++GLV
Sbjct: 375 VQEVVEAEGFRVHPDKTR-----VARKGTRQRVTGLV 406
>PO12_NASVI (Q03270) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 410
Score = 35.0 bits (79), Expect = 2.0
Identities = 18/32 (56%), Positives = 22/32 (68%), Gaps = 1/32 (3%)
Query: 1616 NAKRHRHHLATTASCIRCGAVVEDAEHVLRRC 1647
NA H+ L+ +ASC+ CG V ED EHVL RC
Sbjct: 317 NAFLHKRGLSESASCM-CGDVSEDWEHVLCRC 347
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.336 0.145 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 210,096,723
Number of Sequences: 164201
Number of extensions: 8622404
Number of successful extensions: 27184
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 27096
Number of HSP's gapped (non-prelim): 62
length of query: 1904
length of database: 59,974,054
effective HSP length: 125
effective length of query: 1779
effective length of database: 39,448,929
effective search space: 70179644691
effective search space used: 70179644691
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0228a.4


