

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0227.2
(184 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
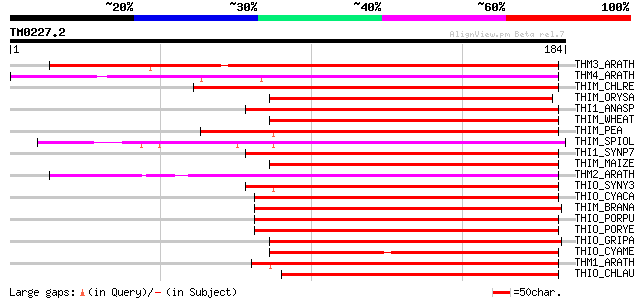
Score E
Sequences producing significant alignments: (bits) Value
THM3_ARATH (Q9SEU7) Thioredoxin M-type 3, chloroplast precursor ... 174 1e-43
THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast precursor ... 121 8e-28
THIM_CHLRE (P23400) Thioredoxin M-type, chloroplast precursor (T... 115 7e-26
THIM_ORYSA (Q9ZP20) Thioredoxin M-type, chloroplast precursor (T... 110 2e-24
THI1_ANASP (P06544) Thioredoxin 1 (TRX-1) (Thioredoxin M) 109 3e-24
THIM_WHEAT (Q9ZP21) Thioredoxin M-type, chloroplast precursor (T... 108 7e-24
THIM_PEA (P48384) Thioredoxin M-type, chloroplast precursor (TRX-M) 108 7e-24
THIM_SPIOL (P07591) Thioredoxin M-type, chloroplast precursor (T... 108 9e-24
THI1_SYNP7 (P12243) Thioredoxin 1 (TRX-1) (Thioredoxin M) 107 2e-23
THIM_MAIZE (Q41864) Thioredoxin M-type, chloroplast precursor (T... 106 3e-23
THM2_ARATH (Q9SEU8) Thioredoxin M-type 2, chloroplast precursor ... 102 5e-22
THIO_SYNY3 (P52231) Thioredoxin (TRX) 101 1e-21
THIO_CYACA (P37395) Thioredoxin 100 2e-21
THIM_BRANA (Q9XGS0) Thioredoxin M-type, chloroplast precursor (T... 98 1e-20
THIO_PORPU (P51225) Thioredoxin 97 2e-20
THIO_PORYE (P50254) Thioredoxin 97 2e-20
THIO_GRIPA (P50338) Thioredoxin 96 5e-20
THIO_CYAME (O22022) Thioredoxin 94 1e-19
THM1_ARATH (O48737) Thioredoxin M-type 1, chloroplast precursor ... 94 2e-19
THIO_CHLAU (Q7M1B9) Thioredoxin (TRX) 91 1e-18
>THM3_ARATH (Q9SEU7) Thioredoxin M-type 3, chloroplast precursor
(TRX-M3)
Length = 173
Score = 174 bits (440), Expect = 1e-43
Identities = 89/170 (52%), Positives = 122/170 (71%), Gaps = 3/170 (1%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSF-PFHLRITHKPLHLQNPPHLPRSHKIRS 72
SSSSSS + + H SS S L P SF P LR + + L + R +
Sbjct: 4 SSSSSSICFNPTRFHTARHISSPSRLFPVTSFSPRSLRFSDRRSLLSSSASRLRLSPL-- 61
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+++RA +T+ WE+S+LKS+ PVLVEFY SWCGPCRMVHR+ID+IA +YAG+LNC+L
Sbjct: 62 CVRDSRAAEVTQRSWEDSVLKSETPVLVEFYTSWCGPCRMVHRIIDEIAGDYAGKLNCYL 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+N D D+ +A++YEIKAVPVV+LFK+G+K ++++GTMPKEFY++AIERVL
Sbjct: 122 LNADNDLPVAEEYEIKAVPVVLLFKNGEKRESIMGTMPKEFYISAIERVL 171
>THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast precursor
(TRX-M4)
Length = 193
Score = 121 bits (304), Expect = 8e-28
Identities = 68/194 (35%), Positives = 109/194 (56%), Gaps = 15/194 (7%)
Query: 1 MASISTSISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQN 60
MAS+ S+++T S ++S SSS S SR ++P+ F + + L Q+
Sbjct: 1 MASLLDSVTVTRVFSLPIAASVSSSSAAP---SVSRRRISPARFLEFRGLKSSRSLVTQS 57
Query: 61 PP-------HLPRSHKIRSLRQETRATPI-----TKDLWENSILKSDIPVLVEFYASWCG 108
+ R +I Q+T A + + W+ +L+SD+PVLVEF+A WCG
Sbjct: 58 ASLGANRRTRIARGGRIACEAQDTTAAAVEVPNLSDSEWQTKVLESDVPVLVEFWAPWCG 117
Query: 109 PCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGT 168
PCRM+H ++D +AK++AG+ + +NTD A+ Y I++VP V++FK G+K D++IG
Sbjct: 118 PCRMIHPIVDQLAKDFAGKFKFYKINTDESPNTANRYGIRSVPTVIIFKGGEKKDSIIGA 177
Query: 169 MPKEFYVNAIERVL 182
+P+E IER L
Sbjct: 178 VPRETLEKTIERFL 191
>THIM_CHLRE (P23400) Thioredoxin M-type, chloroplast precursor
(TRX-M) (Thioredoxin CH2)
Length = 140
Score = 115 bits (287), Expect = 7e-26
Identities = 49/121 (40%), Positives = 78/121 (63%)
Query: 62 PHLPRSHKIRSLRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIA 121
P R+ RS+ A + D ++N +L+S +PVLV+F+A WCGPCR++ V+D+IA
Sbjct: 19 PAFARAAPRRSVVVRAEAGAVNDDTFKNVVLESSVPVLVDFWAPWCGPCRIIAPVVDEIA 78
Query: 122 KEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERV 181
EY +L C +NTD +A +Y I+++P +M+FK G+KC+ +IG +PK V +E+
Sbjct: 79 GEYKDKLKCVKLNTDESPNVASEYGIRSIPTIMVFKGGKKCETIIGAVPKATIVQTVEKY 138
Query: 182 L 182
L
Sbjct: 139 L 139
>THIM_ORYSA (Q9ZP20) Thioredoxin M-type, chloroplast precursor
(TRX-M)
Length = 172
Score = 110 bits (274), Expect = 2e-24
Identities = 46/94 (48%), Positives = 69/94 (72%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
W++ +L S+ PVLVEF+A WCGPCRM+ VID++AKEY G++ C VNTD IA +Y
Sbjct: 75 WDSMVLGSEAPVLVEFWAPWCGPCRMIAPVIDELAKEYVGKIKCCKVNTDDSPNIATNYG 134
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
I+++P V++FK+G+K ++VIG +PK I++
Sbjct: 135 IRSIPTVLMFKNGEKKESVIGAVPKTTLATIIDK 168
>THI1_ANASP (P06544) Thioredoxin 1 (TRX-1) (Thioredoxin M)
Length = 106
Score = 109 bits (273), Expect = 3e-24
Identities = 47/104 (45%), Positives = 71/104 (68%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T ++ +L SD+PVLV+F+A WCGPCRMV V+D+IA++Y G++ VNTD +
Sbjct: 3 AAQVTDSTFKQEVLDSDVPVLVDFWAPWCGPCRMVAPVVDEIAQQYEGKIKVVKVNTDEN 62
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
Q+A Y I+++P +M+FK GQK D V+G +PK +E+ L
Sbjct: 63 PQVASQYGIRSIPTLMIFKGGQKVDMVVGAVPKTTLSQTLEKHL 106
>THIM_WHEAT (Q9ZP21) Thioredoxin M-type, chloroplast precursor
(TRX-M)
Length = 175
Score = 108 bits (270), Expect = 7e-24
Identities = 45/96 (46%), Positives = 68/96 (69%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
W+N ++ + PVLVEF+A WCGPCRM+ VID++AK+Y G++ C VNTD IA Y
Sbjct: 78 WDNMVIACESPVLVEFWAPWCGPCRMIAPVIDELAKDYVGKIKCCKVNTDDCPNIASTYG 137
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
I+++P V++FKDG+K ++VIG +PK I++ +
Sbjct: 138 IRSIPTVLMFKDGEKKESVIGAVPKTTLCTIIDKYI 173
>THIM_PEA (P48384) Thioredoxin M-type, chloroplast precursor (TRX-M)
Length = 172
Score = 108 bits (270), Expect = 7e-24
Identities = 46/120 (38%), Positives = 76/120 (63%), Gaps = 1/120 (0%)
Query: 64 LPRSHKIRSLRQETRATPITKDL-WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAK 122
L +H + R+ + D W+ ++ S+ PVLV+F+A WCGPCRM+ +ID++AK
Sbjct: 53 LIENHVLLHAREAVNEVQVVNDSSWDELVIGSETPVLVDFWAPWCGPCRMIAPIIDELAK 112
Query: 123 EYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
EYAG++ C+ +NTD A Y I+++P V+ FK+G++ D+VIG +PK +E+ +
Sbjct: 113 EYAGKIKCYKLNTDESPNTATKYGIRSIPTVLFFKNGERKDSVIGAVPKATLSEKVEKYI 172
>THIM_SPIOL (P07591) Thioredoxin M-type, chloroplast precursor
(TRX-M)
Length = 181
Score = 108 bits (269), Expect = 9e-24
Identities = 60/184 (32%), Positives = 107/184 (57%), Gaps = 18/184 (9%)
Query: 10 LTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPS--LSFPFH--LRITHKPLHLQNPPHLP 65
L S+S+S + + SH+H HL PS ++ P L+ + L P
Sbjct: 7 LQLSTSASVGTVAVKSHVH---------HLQPSSKVNVPTFRGLKRSFPALSSSVSSSSP 57
Query: 66 RSHKIRSLR-QETRATPITKDL----WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDI 120
R + S+ + + A +D+ W+ +L+S++PV+V+F+A WCGPC+++ VID++
Sbjct: 58 RQFRYSSVVCKASEAVKEVQDVNDSSWKEFVLESEVPVMVDFWAPWCGPCKLIAPVIDEL 117
Query: 121 AKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
AKEY+G++ + +NTD IA Y I+++P V+ FK+G++ +++IG +PK ++IE+
Sbjct: 118 AKEYSGKIAVYKLNTDEAPGIATQYNIRSIPTVLFFKNGERKESIIGAVPKSTLTDSIEK 177
Query: 181 VLKP 184
L P
Sbjct: 178 YLSP 181
>THI1_SYNP7 (P12243) Thioredoxin 1 (TRX-1) (Thioredoxin M)
Length = 106
Score = 107 bits (266), Expect = 2e-23
Identities = 45/104 (43%), Positives = 72/104 (68%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T ++ +L+S IPVLV+F+A WCGPCRMV V+D+IA++Y+ ++ VNTD +
Sbjct: 3 AAAVTDATFKQEVLESSIPVLVDFWAPWCGPCRMVAPVVDEIAQQYSDQVKVVKVNTDEN 62
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+A Y I+++P +M+FKDGQ+ D V+G +PK N +++ L
Sbjct: 63 PSVASQYGIRSIPTLMIFKDGQRVDTVVGAVPKTTLANTLDKHL 106
>THIM_MAIZE (Q41864) Thioredoxin M-type, chloroplast precursor
(TRX-M)
Length = 167
Score = 106 bits (265), Expect = 3e-23
Identities = 43/96 (44%), Positives = 67/96 (69%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
W+ ++ + PVLVEF+A WCGPCRM+ VID++AK+YAG++ C VNTD +A Y
Sbjct: 69 WDGLVMACETPVLVEFWAPWCGPCRMIAPVIDELAKDYAGKITCCKVNTDDSPNVASTYG 128
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
I+++P V++FK G+K ++VIG +PK I++ +
Sbjct: 129 IRSIPTVLIFKGGEKKESVIGAVPKSTLTTLIDKYI 164
>THM2_ARATH (Q9SEU8) Thioredoxin M-type 2, chloroplast precursor
(TRX-M2)
Length = 186
Score = 102 bits (254), Expect = 5e-22
Identities = 53/169 (31%), Positives = 92/169 (54%), Gaps = 5/169 (2%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQNPPHLPRSHKIRSL 73
SSS S+SS SS + SS + SLS P L H+P L R+ +
Sbjct: 22 SSSPSASSLSSRRMFAVLPESSGLRIRLSLS-PASLTSIHQP----RVSRLRRAVVCEAQ 76
Query: 74 RQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLV 133
T + W++ +LK+ PV+V+F+A WCGPC+M+ +++D+A+ Y G++ + +
Sbjct: 77 ETTTDIQVVNDSTWDSLVLKATGPVVVDFWAPWCGPCKMIDPLVNDLAQHYTGKIKFYKL 136
Query: 134 NTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
NTD Y ++++P +M+F G+K D +IG +PK +++++ L
Sbjct: 137 NTDESPNTPGQYGVRSIPTIMIFVGGEKKDTIIGAVPKTTLTSSLDKFL 185
>THIO_SYNY3 (P52231) Thioredoxin (TRX)
Length = 106
Score = 101 bits (251), Expect = 1e-21
Identities = 44/105 (41%), Positives = 71/105 (66%), Gaps = 1/105 (0%)
Query: 79 ATPITKDL-WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDT 137
ATP D ++ +L S++PVLV+F+A WCGPCRMV V+D+I+++Y G++ +NTD
Sbjct: 2 ATPQVSDASFKEDVLDSELPVLVDFWAPWCGPCRMVAPVVDEISQQYEGKVKVVKLNTDE 61
Query: 138 DMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+ A Y I+++P +M+FK GQ+ D V+G +PK + +E+ L
Sbjct: 62 NPNTASQYGIRSIPTLMIFKGGQRVDMVVGAVPKTTLASTLEKYL 106
>THIO_CYACA (P37395) Thioredoxin
Length = 107
Score = 100 bits (248), Expect = 2e-21
Identities = 43/101 (42%), Positives = 70/101 (68%)
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+T +E ++ S+ VLV+F+A WCGPCRM+ VID++A+EY ++ +NTD + I
Sbjct: 7 VTDFSFEKEVVNSEKLVLVDFWAPWCGPCRMISPVIDELAQEYVEQVKIVKINTDENPSI 66
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+ +Y I+++P +MLFKDG++ D VIG +PK NA+++ L
Sbjct: 67 SAEYGIRSIPTLMLFKDGKRVDTVIGAVPKSTLTNALKKYL 107
>THIM_BRANA (Q9XGS0) Thioredoxin M-type, chloroplast precursor
(TRX-M)
Length = 177
Score = 97.8 bits (242), Expect = 1e-20
Identities = 36/102 (35%), Positives = 68/102 (66%)
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+ WE+ +LK+D PV+V+F+A WCGPC+M+ +++++A++Y G++ F +NTD
Sbjct: 76 VNDSTWESLVLKADEPVVVDFWAPWCGPCKMIDPIVNELAQQYTGKIKFFKLNTDDSPAT 135
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLK 183
Y ++++P +M+F G+K D +IG +PK +I++ L+
Sbjct: 136 PGKYGVRSIPTIMIFVKGEKKDTIIGAVPKTTLATSIDKFLQ 177
>THIO_PORPU (P51225) Thioredoxin
Length = 107
Score = 97.4 bits (241), Expect = 2e-20
Identities = 40/101 (39%), Positives = 67/101 (65%)
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+T ++ ++ +D+PVLV+F+A WCGPCRMV V+D IA+EY + +NTD + I
Sbjct: 6 VTDASFKQEVINNDLPVLVDFWAPWCGPCRMVSPVVDAIAEEYESSIKVVKINTDDNPTI 65
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
A +Y I+++P +M+FK G++ D VIG +PK + + + +
Sbjct: 66 AAEYGIRSIPTLMIFKSGERVDTVIGAVPKSTLESTLNKYI 106
>THIO_PORYE (P50254) Thioredoxin
Length = 107
Score = 97.1 bits (240), Expect = 2e-20
Identities = 39/101 (38%), Positives = 68/101 (66%)
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+T ++ ++ +++PVLV+F+A WCGPCRMV V+D+IA+EY + +NTD + I
Sbjct: 6 VTDASFKQEVINNNLPVLVDFWAPWCGPCRMVSPVVDEIAEEYESSIKVVKINTDDNPTI 65
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
A +Y I+++P +M+FK G++ D VIG +PK + + + +
Sbjct: 66 AAEYGIRSIPTLMIFKAGERVDTVIGAVPKSTLASTLNKYI 106
>THIO_GRIPA (P50338) Thioredoxin
Length = 109
Score = 95.9 bits (237), Expect = 5e-20
Identities = 42/97 (43%), Positives = 65/97 (66%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
+ ++ S PVLV+F+A WCGPCRM+ ID+IA +Y +L VNTD + IA +Y
Sbjct: 11 FHEEVINSRQPVLVDFWAPWCGPCRMIASTIDEIAHDYKDKLKVVKVNTDQNPTIATEYG 70
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLK 183
I+++P VM+F +G+K D V+G +PK +N +++ LK
Sbjct: 71 IRSIPTVMIFINGKKVDTVVGAVPKLTLLNTLQKHLK 107
>THIO_CYAME (O22022) Thioredoxin
Length = 102
Score = 94.4 bits (233), Expect = 1e-19
Identities = 43/96 (44%), Positives = 66/96 (67%), Gaps = 2/96 (2%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
+EN +L+S+ VLV+F+A WCGPCRM+ ++++IAKE+ L VNTD + +A Y
Sbjct: 9 FENEVLQSEKLVLVDFWAPWCGPCRMIGPILEEIAKEF--NLKVVQVNTDENPNLATFYG 66
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
I+++P +MLFK GQ+ D VIG +PK ++ I + L
Sbjct: 67 IRSIPTLMLFKKGQRVDTVIGAVPKSILIHTINKYL 102
>THM1_ARATH (O48737) Thioredoxin M-type 1, chloroplast precursor
(TRX-M1)
Length = 179
Score = 94.0 bits (232), Expect = 2e-19
Identities = 36/103 (34%), Positives = 68/103 (65%), Gaps = 1/103 (0%)
Query: 81 PITKD-LWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDM 139
P+ D W++ +LK+D PV V+F+A WCGPC+M+ +++++A++YAG+ + +NTD
Sbjct: 77 PVVNDSTWDSLVLKADEPVFVDFWAPWCGPCKMIDPIVNELAQKYAGQFKFYKLNTDESP 136
Query: 140 QIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
Y ++++P +M+F +G+K D +IG + K+ +I + L
Sbjct: 137 ATPGQYGVRSIPTIMIFVNGEKKDTIIGAVSKDTLATSINKFL 179
>THIO_CHLAU (Q7M1B9) Thioredoxin (TRX)
Length = 109
Score = 91.3 bits (225), Expect = 1e-18
Identities = 38/92 (41%), Positives = 62/92 (67%)
Query: 91 ILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAV 150
+L+S PV+V+F+A WCGPCR++ ++D +A EYAGRL VNTD ++Q A +K +
Sbjct: 15 VLQSKTPVVVDFWAPWCGPCRVIAPILDKLAGEYAGRLTIAKVNTDDNVQYASQLGLKGL 74
Query: 151 PVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
P +++FKDG++ ++G P+ Y ++VL
Sbjct: 75 PTLVIFKDGREVGRLVGARPEAMYREIFDKVL 106
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.131 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,003,704
Number of Sequences: 164201
Number of extensions: 857762
Number of successful extensions: 11561
Number of sequences better than 10.0: 416
Number of HSP's better than 10.0 without gapping: 348
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 7252
Number of HSP's gapped (non-prelim): 1918
length of query: 184
length of database: 59,974,054
effective HSP length: 104
effective length of query: 80
effective length of database: 42,897,150
effective search space: 3431772000
effective search space used: 3431772000
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0227.2


