

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
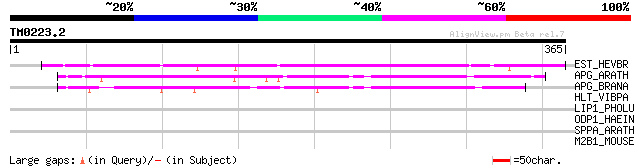
Query= TM0223.2
(365 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early nodule... 272 8e-73
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 96 1e-19
APG_BRANA (P40603) Anter-specific proline-rich protein APG (Prot... 96 2e-19
HLT_VIBPA (Q99289) Thermolabile hemolysin precursor (TL) (Lecith... 42 0.002
LIP1_PHOLU (P40601) Lipase 1 precursor (EC 3.1.1.3) (Triacylglyc... 39 0.021
ODP1_HAEIN (P45119) Pyruvate dehydrogenase E1 component (EC 1.2.... 32 2.5
SPPA_ARATH (Q9SEL7) Protease sppA, chloroplast precursor (EC 3.4... 31 4.3
M2B1_MOUSE (O09159) Lysosomal alpha-mannosidase precursor (EC 3.... 30 9.6
>EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early
nodule-specific protein homolog) (Latex allergen Hev b
13)
Length = 391
Score = 272 bits (696), Expect = 8e-73
Identities = 157/367 (42%), Positives = 207/367 (55%), Gaps = 30/367 (8%)
Query: 22 SAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVID 81
S A CD PA+ F FGDSNSDTGG + +P+N P G TFFHRSTGR SDGRL+ID
Sbjct: 22 SLAYASETCDFPAI-FNFGDSNSDTGGKAAAF-YPLNPPYGETFFHRSTGRYSDGRLIID 79
Query: 82 LLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSST-LPKYV--------PFSLNIQVMQ 132
+ +S N +L+PYL SL GS+F +GA+FA GS+ LP + PF L++Q Q
Sbjct: 80 FIAESFNLPYLSPYLSSL-GSNFKHGADFATAGSTIKLPTTIIPAHGGFSPFYLDVQYSQ 138
Query: 133 FLHFKARTLELVSAG---AKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIP 189
F F R+ + G A+ V + F ALY DIGQNDL + F NLT +V +P
Sbjct: 139 FRQFIPRSQFIRETGGIFAELVPEEYYFEKALYTFDIGQNDLTEGFL-NLTVEEVNATVP 197
Query: 190 TVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSYNSAARLF 249
++ VK +Y+ GAR FW+HNTGP+GCL IL + D GC +YN A+ F
Sbjct: 198 DLVNSFSANVKKIYDLGARTFWIHNTGPIGCLSFILTYFPWAEKDSAGCAKAYNEVAQHF 257
Query: 250 NEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDL 309
N L +LR L AT V+VDIY++K+ L + K+GF PL CCG+GG YNF +
Sbjct: 258 NHKLKEIVAQLRKDLPLATFVHVDIYSVKYSLFSEPEKHGFEFPLITCCGYGG-KYNFSV 316
Query: 310 RVTCGQPGYQVCDEGSRY-----------VNWDGTHHTEAANTFIASKILSTDYSTPRTP 358
CG D+G++ VNWDG H+TEAAN + +I + +S P P
Sbjct: 317 TAPCGDT--VTADDGTKIVVGSCACPSVRVNWDGAHYTEAANEYFFDQISTGAFSDPPVP 374
Query: 359 FEFFCHR 365
CH+
Sbjct: 375 LNMACHK 381
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 96.3 bits (238), Expect = 1e-19
Identities = 101/342 (29%), Positives = 147/342 (42%), Gaps = 36/342 (10%)
Query: 32 VPAVLFVFGDSNSDTGGLTSGLGFPINL---PNGRTF-FHRSTGRLSDGRLVIDLLCQSL 87
+PAV F FGDS DTG + L I P G F F +TGR S+G + D L + +
Sbjct: 202 IPAVFF-FGDSVFDTGN-NNNLETKIKSNYRPYGMDFKFRVATGRFSNGMVASDYLAKYM 259
Query: 88 NTRFLTP-YLD-SLSGSSFTNGANFAVVGSSTLPKYVPFSLNIQVMQFLHFKARTLELVS 145
+ + P YLD + + G +FA G+ P + I ++ L + +E V+
Sbjct: 260 GVKEIVPAYLDPKIQPNDLLTGVSFASGGAGYNPTTSEAANAIPMLDQLTYFQDYIEKVN 319
Query: 146 A---GAKNVINDEGFRAALYLIDIG-------QNDLADSF----AKNLTYVQVIKKIPTV 191
K+ G LI G NDL ++ A+ L I T+
Sbjct: 320 RLVRQEKSQYKLAGLEKTNQLISKGVAIVVGGSNDLIITYFGSGAQRLK--NDIDSYTTI 377
Query: 192 ITE-IENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSYNSAARLFN 250
I + + V LY GAR+ V T PLGC+P +KK + C N A++LFN
Sbjct: 378 IADSAASFVLQLYGYGARRIGVIGTPPLGCVPS--QRLKKKKI----CNEELNYASQLFN 431
Query: 251 EALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLR 310
L +L L ++T VY+DIY I ++ YGF CC G
Sbjct: 432 SKLLLILGQLSKTLPNSTFVYMDIYTIISQMLETPAAYGFEETKKPCCKTG----LLSAG 487
Query: 311 VTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDY 352
C + ++C S Y+ WDG H T+ A I +K+L +Y
Sbjct: 488 ALCKKSTSKICPNTSSYLFWDGVHPTQRAYKTI-NKVLIKEY 528
>APG_BRANA (P40603) Anter-specific proline-rich protein APG (Protein
CEX) (Fragment)
Length = 449
Score = 95.5 bits (236), Expect = 2e-19
Identities = 95/337 (28%), Positives = 146/337 (43%), Gaps = 55/337 (16%)
Query: 32 VPAVLFVFGDSNSDTGGLTS------------GLGFPINLPNGRTFFHRSTGRLSDGRLV 79
+PAV F FGDS DTG + G+ FP+ + +TGR S+GR+
Sbjct: 123 IPAVFF-FGDSIFDTGNNNNLDTKLKCNYRPYGMDFPMGV---------ATGRFSNGRVA 172
Query: 80 IDLLCQSLNTRFLTP-YLDS-------LSGSSFTNGANFAVVGSSTLPK----YVPFSLN 127
D + + L + + P Y+D L S G +FA G+ LP+ + ++
Sbjct: 173 SDYISKYLGVKEIVPAYVDKKLQQNNELQQSDLLTGVSFASGGAGYLPQTSESWKVTTML 232
Query: 128 IQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKK 187
Q+ F +K R +LV I +G ++ G NDL ++ N Q +K
Sbjct: 233 DQLTYFQDYKKRMKKLVGKKKTKKIVSKGAA----IVVAGSNDLIYTYFGN--GAQHLKN 286
Query: 188 -IPTVITEIENAVKS----LYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSY 242
+ + T + ++ S LY GAR+ V T P+GC P +KK + C
Sbjct: 287 DVDSFTTMMADSAASFVLQLYGYGARRIGVIGTPPIGCTPS--QRVKKKKI----CNEDL 340
Query: 243 NSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGG 302
N AA+LFN L +L L ++T+VY DIY+I ++ + YGF CC G
Sbjct: 341 NYAAQLFNSKLVIILGQLSKTLPNSTIVYGDIYSIFSKMLESPEDYGFEEIKKPCCKIGL 400
Query: 303 PPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAA 339
V C + + S Y+ WDG H ++ A
Sbjct: 401 TKGG----VFCKERTLKNMSNASSYLFWDGLHPSQRA 433
>HLT_VIBPA (Q99289) Thermolabile hemolysin precursor (TL)
(Lecithin-dependent haemolysin) (LDH) (Atypical
phospholipase) (Phospholipase A2) (Lysophospholipase)
Length = 418
Score = 42.0 bits (97), Expect = 0.002
Identities = 68/339 (20%), Positives = 127/339 (37%), Gaps = 67/339 (19%)
Query: 18 ITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGR 77
I S+ AA+ + + L GDS SDTG + + + PN ++F G S+G
Sbjct: 132 IWSNDAAMQPDQINKVVAL---GDSLSDTGNIFNASQW--RFPNPNSWF---LGHFSNGF 183
Query: 78 LVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVPFS-LNIQVMQFLHF 136
+ + + ++ N N+AV G++ +Y+ + + QV +L +
Sbjct: 184 VWTEYIAKAKNLPLY----------------NWAVGGAAGENQYIALTGVGEQVSSYLTY 227
Query: 137 KARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIE 196
A L+ ++ G ND ++ + +P V +
Sbjct: 228 AKLAKNYKPANT------------LFTLEFGLND----------FMNYNRGVPEVKADYA 265
Query: 197 NAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSYNSAARLFNEALYHS 256
A+ L + GA+ F + P+ Q++ + + N + +A+Y+
Sbjct: 266 EALIRLTDAGAKNFMLMTLPDATKAPQFKYSTQEEIDKIRAKVLEMNEFIKA--QAMYYK 323
Query: 257 SQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPP-----YNFDLRV 311
+Q L D +A+ L + ++GF N C Y LR
Sbjct: 324 AQGYNITL-------FDTHALFETLTSAPEEHGFVNASDPCLDINRSSSVDYMYTHALRS 376
Query: 312 TCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILST 350
C G + ++V WD TH T A + ++A K+L +
Sbjct: 377 ECAASGAE------KFVFWDVTHPTTATHRYVAEKMLES 409
>LIP1_PHOLU (P40601) Lipase 1 precursor (EC 3.1.1.3)
(Triacylglycerol lipase)
Length = 645
Score = 38.9 bits (89), Expect = 0.021
Identities = 74/320 (23%), Positives = 115/320 (35%), Gaps = 97/320 (30%)
Query: 36 LFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPY 95
L+VFGDS SD G NGR G ++ +L D + Q L +
Sbjct: 28 LYVFGDSLSDGGN------------NGRYTVDGING--TESKLYNDFIAQQLGIELV--- 70
Query: 96 LDSLSGSSFTNGANFAVVGSSTLPKYV-PFSLNIQVMQFLHFKARTLELVSAGAKNVIND 154
+S G N+A G++ + + QVM +L A N +
Sbjct: 71 ------NSKKGGTNYAAGGATAVADLNNKHNTQDQVMGYL-----------ASHSNRADH 113
Query: 155 EGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHN 214
G +Y+ IG ND+ D+ +N Q I I + V +L N GA V
Sbjct: 114 NG----MYVHWIGGNDV-DAALRNPADAQKI--ITESAMAASSQVHALLNAGAGLVIVPT 166
Query: 215 TGPLGCLPKILAL------AQKKDL-------------------------------DLFG 237
+G PKI+ A KDL D+ G
Sbjct: 167 VPDVGMTPKIMEFVLSKGGATSKDLAKIHAVVNGYPTIDKDTRLQVIHGVFKQIGSDVSG 226
Query: 238 ------------CLSSYN----SAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDL 281
+ YN +A++L + Y+ + + ++ +V VD+ A+ H++
Sbjct: 227 GDAKKAEETTKQLIDGYNELSSNASKLVDN--YNQLEDMALSQENGNIVRVDVNALLHEV 284
Query: 282 IANATKYGFSNPLTVCCGFG 301
IAN +YGF N + C G
Sbjct: 285 IANPLRYGFLNTIGYACAQG 304
>ODP1_HAEIN (P45119) Pyruvate dehydrogenase E1 component (EC
1.2.4.1)
Length = 886
Score = 32.0 bits (71), Expect = 2.5
Identities = 18/49 (36%), Positives = 28/49 (56%), Gaps = 1/49 (2%)
Query: 228 AQKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYA 276
+QKKDLDL G +S++ SAA ++ E Y+ K T L++ +A
Sbjct: 96 SQKKDLDLGGHISTFQSAATMY-EVCYNHFFKAATEKNGGDLIFFQGHA 143
>SPPA_ARATH (Q9SEL7) Protease sppA, chloroplast precursor (EC
3.4.21.-)
Length = 321
Score = 31.2 bits (69), Expect = 4.3
Identities = 25/88 (28%), Positives = 43/88 (48%), Gaps = 3/88 (3%)
Query: 39 FGDSNSDTGGLTSGLGFPINLPNGRTFFH--RSTGRLSDGRLVIDLLCQSLNTRFLTPYL 96
+G N+ T G+ SGLG I PNG++ ++ ++ G LL +T +
Sbjct: 223 YGYENTLTIGVVSGLGREIPSPNGKSISEAIQTDADINSGNSGGPLLDSYGHTIGVNTAT 282
Query: 97 DSLSGSSFTNGANFAVVGSSTLPKYVPF 124
+ GS ++G NFA + T+ + VP+
Sbjct: 283 FTRKGSGMSSGVNFA-IPIDTVVRTVPY 309
>M2B1_MOUSE (O09159) Lysosomal alpha-mannosidase precursor (EC
3.2.1.24) (Mannosidase, alpha B) (Lysosomal acid
alpha-mannosidase) (Laman) (Mannosidase alpha class 2B
member 1)
Length = 1013
Score = 30.0 bits (66), Expect = 9.6
Identities = 23/108 (21%), Positives = 46/108 (42%), Gaps = 7/108 (6%)
Query: 114 GSSTLPKYVPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLAD 173
G T P P LN+ ++ H L+ V +++D + Y++D + L +
Sbjct: 51 GYKTCPPTKPGMLNVHLLPHTHDDVGWLKTVDQYYYGILSDVQHASVQYILDSVVSSLLE 110
Query: 174 SFAKNLTYVQVI---KKIPTVITEIENAVKSLYNEGARKF----WVHN 214
+ YV++ + + ++AV++L +G +F WV N
Sbjct: 111 KPTRRFIYVEMAFFSRWWKQQTSATQDAVRNLVRQGRLEFVNGGWVMN 158
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,168,498
Number of Sequences: 164201
Number of extensions: 1845554
Number of successful extensions: 3576
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 3559
Number of HSP's gapped (non-prelim): 9
length of query: 365
length of database: 59,974,054
effective HSP length: 112
effective length of query: 253
effective length of database: 41,583,542
effective search space: 10520636126
effective search space used: 10520636126
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0223.2


