

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
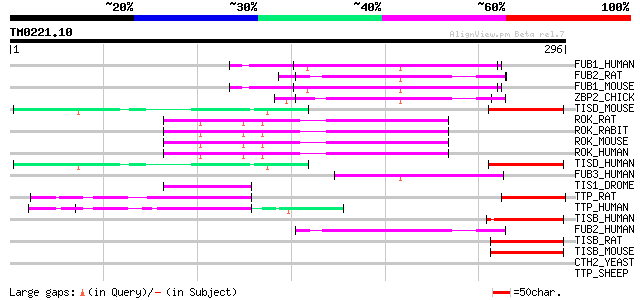
Query= TM0221.10
(296 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
FUB1_HUMAN (Q96AE4) Far upstream element binding protein 1 (FUSE... 53 8e-07
FUB2_RAT (Q99PF5) Far upstream element binding protein 2 (FUSE b... 52 1e-06
FUB1_MOUSE (Q91WJ8) Far upstream element binding protein 1 (FUSE... 52 1e-06
ZBP2_CHICK (Q8UVD9) Zipcode-binding protein 2 50 9e-06
TISD_MOUSE (P23949) Butyrate response factor 2 (TIS11D protein) 49 1e-05
ROK_RAT (P61980) Heterogeneous nuclear ribonucleoprotein K (dC-s... 49 1e-05
ROK_RABIT (O19049) Heterogeneous nuclear ribonucleoprotein K (hn... 49 1e-05
ROK_MOUSE (P61979) Heterogeneous nuclear ribonucleoprotein K 49 1e-05
ROK_HUMAN (P61978) Heterogeneous nuclear ribonucleoprotein K (hn... 49 1e-05
TISD_HUMAN (P47974) Butyrate response factor 2 (TIS11D protein) ... 49 1e-05
FUB3_HUMAN (Q96I24) Far upstream element binding protein 3 (FUSE... 49 1e-05
TIS1_DROME (P47980) TIS11 protein (dTIS11) 47 4e-05
TTP_RAT (P47973) Tristetraproline (TTP) (Zinc finger protein 36)... 44 4e-04
TTP_HUMAN (P26651) Tristetraproline (TTP) (Zinc finger protein 3... 44 4e-04
TISB_HUMAN (Q07352) Butyrate response factor 1 (TIS11B protein) ... 44 4e-04
FUB2_HUMAN (Q92945) Far upstream element binding protein 2 (FUSE... 44 5e-04
TISB_RAT (P17431) Butyrate response factor 1 (TIS11B protein) (E... 44 6e-04
TISB_MOUSE (P23950) Butyrate response factor 1 (TIS11B protein) 44 6e-04
CTH2_YEAST (P47977) Zinc finger protein CTH2 (YTIS11 protein) 43 0.001
TTP_SHEEP (Q6S9E0) Tristetraproline (TTP) (Zinc finger protein 3... 42 0.001
>FUB1_HUMAN (Q96AE4) Far upstream element binding protein 1 (FUSE
binding protein 1) (FBP) (DNA helicase V) (HDH V)
Length = 643
Score = 53.1 bits (126), Expect = 8e-07
Identities = 44/153 (28%), Positives = 65/153 (41%), Gaps = 17/153 (11%)
Query: 118 DKCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEP-----PPGPATGFGANATAK 172
D+C A E+ + S P G P GR GR + PPG F
Sbjct: 329 DRCQHAA---EIITDLLRSVQAGNPGGPGPGGRGRGRGQGNWNMGPPGGLQEFN------ 379
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREH---ESDPNLRNIELEGSFDQIKEAS 229
V G IIGKGG K I +Q+GA++ ++ + +DPN++ + G+ QI A
Sbjct: 380 FIVPTGKTGLIIGKGGETIKSISQQSGARIELQRNPPPNADPNMKLFTIRGTPQQIDYAR 439
Query: 230 NMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHG 262
++++ + P G G PG HG G
Sbjct: 440 QLIEEKIGGPVNPLGPPVPHGPHGVPGPHGPPG 472
Score = 43.5 bits (101), Expect = 6e-04
Identities = 26/109 (23%), Positives = 46/109 (41%)
Query: 152 GGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDP 211
GG E + G N + + G +IG+ G K+I G ++ + +
Sbjct: 258 GGFREVRNEYGSRIGGNEGIDVPIPRFAVGIVIGRNGEMIKKIQNDAGVRIQFKPDDGTT 317
Query: 212 NLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGH 260
R ++ G D+ + A+ ++ DLL ++Q P GG G G G+
Sbjct: 318 PERIAQITGPPDRCQHAAEIITDLLRSVQAGNPGGPGPGGRGRGRGQGN 366
Score = 40.4 bits (93), Expect = 0.005
Identities = 26/83 (31%), Positives = 45/83 (53%), Gaps = 3/83 (3%)
Query: 157 PPPGPATGFG-ANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREH--ESDPNL 213
P PG G G NA +I + AS AG +IGKGG KQ+ + G K+ + + ++
Sbjct: 172 PAPGFHHGDGPGNAVQEIMIPASKAGLVIGKGGETIKQLQERAGVKMVMIQDGPQNTGAD 231
Query: 214 RNIELEGSFDQIKEASNMVKDLL 236
+ + + G ++++A MV +L+
Sbjct: 232 KPLRITGDPYKVQQAKEMVLELI 254
Score = 33.1 bits (74), Expect = 0.85
Identities = 22/96 (22%), Positives = 40/96 (40%), Gaps = 12/96 (12%)
Query: 170 TAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEAS 229
T + V + G IIG+GG +I +++G K+ I R+ L G+ + ++ A
Sbjct: 101 TEEYKVPDGMVGFIIGRGGEQISRIQQESGCKIQIAPDSGGLPERSCMLTGTPESVQSAK 160
Query: 230 NMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNN 265
++ ++ + G PG H G N
Sbjct: 161 RLLDQIV------------EKGRPAPGFHHGDGPGN 184
>FUB2_RAT (Q99PF5) Far upstream element binding protein 2 (FUSE
binding protein 2) (KH type splicing regulatory protein)
(KSRP) (MAP2 RNA trans-acting protein 1) (MARTA1)
Length = 721
Score = 52.4 bits (124), Expect = 1e-06
Identities = 39/125 (31%), Positives = 52/125 (41%), Gaps = 8/125 (6%)
Query: 144 GHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLS 203
G P GR GR + GP G S+ G +IG+GG N K I +QTGA +
Sbjct: 406 GMPPGGRGRGRGQGNWGPPGG-----EMTFSIPTHKCGLVIGRGGENVKAINQQTGAFVE 460
Query: 204 IREH---ESDPNLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGH 260
I DPN + + GS QI A ++++ + P G GP G H
Sbjct: 461 ISRQLPPNGDPNFKLFVIRGSPQQIDHAKQLIEEKIEGPLCPVGPGPGGPGPAGPMGPFH 520
Query: 261 HGSNN 265
G N
Sbjct: 521 PGPFN 525
Score = 43.9 bits (102), Expect = 5e-04
Identities = 29/112 (25%), Positives = 49/112 (42%), Gaps = 18/112 (16%)
Query: 153 GRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPN 212
G + PPP + T + V + G IIG+GG +I + +G K+ I
Sbjct: 136 GPIHPPPR------TSMTEEYRVPDGMVGLIIGRGGEQINKIQQDSGCKVQISPDSGGLP 189
Query: 213 LRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSN 264
R++ L G+ + +++A M+ D++ G GGP G H +N
Sbjct: 190 ERSVSLTGAPESVQKAKMMLDDIV------------SRGRGGPPGQFHDNAN 229
Score = 40.0 bits (92), Expect = 0.007
Identities = 30/106 (28%), Positives = 50/106 (46%), Gaps = 6/106 (5%)
Query: 158 PPGP----ATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAK-LSIREHESDPN 212
PPG A G +I + A AG +IGKGG KQ+ + G K + I++ + N
Sbjct: 220 PPGQFHDNANGGQNGTVQEIMIPAGKAGLVIGKGGETIKQLQERAGVKMILIQDGSQNTN 279
Query: 213 L-RNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGG 257
+ + + + G ++++A MV D+L N+ G+ GG
Sbjct: 280 VDKPLRIIGDPYKVQQACEMVMDILRERDQGGFGDRNEYGSRVGGG 325
>FUB1_MOUSE (Q91WJ8) Far upstream element binding protein 1 (FUSE
binding protein 1) (FBP)
Length = 651
Score = 52.4 bits (124), Expect = 1e-06
Identities = 44/153 (28%), Positives = 64/153 (41%), Gaps = 17/153 (11%)
Query: 118 DKCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEP-----PPGPATGFGANATAK 172
D+C A E+ + S P G P GR GR + PPG F
Sbjct: 326 DRCQHAA---EIITDLLRSVQAGNPGGPGPGGRGRGRGQGNWNMGPPGGLQEFN------ 376
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREH---ESDPNLRNIELEGSFDQIKEAS 229
V G IIGKGG K I +Q+GA++ ++ +DPN++ + G+ QI A
Sbjct: 377 FIVPTGKTGLIIGKGGETIKSISQQSGARIELQRSPPPNADPNMKLFTIRGTPQQIDYAR 436
Query: 230 NMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHG 262
++++ + P G G PG HG G
Sbjct: 437 QLIEEKIGGPVNPLGPPVPHGPHGVPGPHGPPG 469
Score = 43.1 bits (100), Expect = 8e-04
Identities = 26/109 (23%), Positives = 46/109 (41%)
Query: 152 GGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDP 211
GG E + G N + + G +IG+ G K+I G ++ + +
Sbjct: 255 GGFREVRNEYGSRIGGNEGIDVPIPRFAVGIVIGRNGEMIKKIQNDAGVRIQFKPDDGTT 314
Query: 212 NLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGH 260
R ++ G D+ + A+ ++ DLL ++Q P GG G G G+
Sbjct: 315 PDRIAQITGPPDRCQHAAEIITDLLRSVQAGNPGGPGPGGRGRGRGQGN 363
Score = 40.4 bits (93), Expect = 0.005
Identities = 26/83 (31%), Positives = 45/83 (53%), Gaps = 3/83 (3%)
Query: 157 PPPGPATGFG-ANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREH--ESDPNL 213
P PG G G NA +I + AS AG +IGKGG KQ+ + G K+ + + ++
Sbjct: 169 PAPGFHHGDGPGNAVQEIMIPASKAGLVIGKGGETIKQLQERAGVKMVMIQDGPQNTGAD 228
Query: 214 RNIELEGSFDQIKEASNMVKDLL 236
+ + + G ++++A MV +L+
Sbjct: 229 KPLRITGDPYKVQQAKEMVLELI 251
Score = 33.1 bits (74), Expect = 0.85
Identities = 22/96 (22%), Positives = 40/96 (40%), Gaps = 12/96 (12%)
Query: 170 TAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEAS 229
T + V + G IIG+GG +I +++G K+ I R+ L G+ + ++ A
Sbjct: 98 TEEYKVPDGMVGFIIGRGGEQISRIQQESGCKIQIAPDSGGLPERSCMLTGTPESVQSAK 157
Query: 230 NMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNN 265
++ ++ + G PG H G N
Sbjct: 158 RLLDQIV------------EKGRPAPGFHHGDGPGN 181
>ZBP2_CHICK (Q8UVD9) Zipcode-binding protein 2
Length = 769
Score = 49.7 bits (117), Expect = 9e-06
Identities = 38/121 (31%), Positives = 51/121 (41%), Gaps = 10/121 (8%)
Query: 142 PIGHA--PAGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTG 199
P GH P GR GR + GP G S+ G +IG+GG N K I +Q G
Sbjct: 463 PPGHGMPPGGRGRGRGQGIWGPPGG-----EMTFSIPTHKCGLVIGRGGENVKAINQQRG 517
Query: 200 AKLSIREH---ESDPNLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPG 256
A + I DPN + + GS QI+ A +++ + P GG GP
Sbjct: 518 AFVEISRQLPPNGDPNFKLFIIRGSPQQIEHAKQPIEEKIEGPLCPVGPGPGPGGPPGPA 577
Query: 257 G 257
G
Sbjct: 578 G 578
Score = 46.6 bits (109), Expect = 7e-05
Identities = 32/112 (28%), Positives = 49/112 (43%), Gaps = 17/112 (15%)
Query: 153 GRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPN 212
G M PPP T T + V + G IIG+GG +I + +G K+ I
Sbjct: 198 GPMHPPPRSTT-----VTEEYRVPDGMVGLIIGRGGEQINKIQQDSGCKVQISPDSGGLP 252
Query: 213 LRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSN 264
R++ L GS + +++A M+ D++ G GGP G H +N
Sbjct: 253 ERSVSLTGSPEAVQKAKLMLDDIV------------SRGRGGPPGQFHDYAN 292
Score = 42.0 bits (97), Expect = 0.002
Identities = 56/211 (26%), Positives = 84/211 (39%), Gaps = 45/211 (21%)
Query: 76 PSAQAPPRNVAAPPPP----------VPNGSTPAVKTR---ICNKFNTAEGCK------- 115
PSA PP+ PPP VP+G + R NK GCK
Sbjct: 189 PSAALPPQLGPMHPPPRSTTVTEEYRVPDGMVGLIIGRGGEQINKIQQDSGCKVQISPDS 248
Query: 116 --FGDKCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPATGF--GANATA 171
++ G E + DD G GG PPG + G N T
Sbjct: 249 GGLPERSVSLTGSPEAVQKAKLMLDDIVSRGR------GG----PPGQFHDYANGQNGTV 298
Query: 172 K-ISVEASLAGAIIGKGGVNSKQICRQTGAK-LSIREHESDPNL-RNIELEGSFDQIKEA 228
+ I + A AG +IGKGG KQ+ + G K + I++ + N+ + + + G ++++A
Sbjct: 299 QEIMIPAGKAGLVIGKGGETIKQLQERAGVKMIFIQDGSQNTNVDKPLRIIGDPYKVQQA 358
Query: 229 SNMVKDLLLTLQMSAPPKSNQGGAGGPGGHG 259
MV D+L + +QGG G +G
Sbjct: 359 CEMVMDIL--------RERDQGGFGDRNEYG 381
Score = 36.6 bits (83), Expect = 0.077
Identities = 22/87 (25%), Positives = 41/87 (46%), Gaps = 9/87 (10%)
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMV 232
+ V G +IG+ G K+I G ++ ++ + + + G ++ + A+ ++
Sbjct: 390 VPVPRHSVGVVIGRSGEMIKKIQNDAGVRIQFKQDDGTGPEKIAHIMGPPERCEHAARII 449
Query: 233 KDLLLTLQMSAPPKSNQGGAGGPGGHG 259
DLL +L+ S PP GP GHG
Sbjct: 450 NDLLQSLR-SGPP--------GPPGHG 467
>TISD_MOUSE (P23949) Butyrate response factor 2 (TIS11D protein)
Length = 367
Score = 49.3 bits (116), Expect = 1e-05
Identities = 43/165 (26%), Positives = 61/165 (36%), Gaps = 37/165 (22%)
Query: 3 FRKRGRPEPGFNSNGGIKKSKQELESLSSGVGS---KSKPCTKFFSTAGCPFGESCHFLH 59
FR R E G S + +Q+ S + S K++ C F + C +GE C F H
Sbjct: 92 FRDRSFSENGERSQHLLHLQQQQKGGSGSQINSTRYKTELCRPFEESGTCKYGEKCQFAH 151
Query: 60 YVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDK 119
G++ + + P KT +C F+T C +G +
Sbjct: 152 ----GFHELRSLTR-----------------------HPKYKTELCRTFHTIGFCPYGPR 184
Query: 120 CHFAHGEWELGKHIAPS-----FDDHRPIGHAPAGRIGGRMEPPP 159
CHF H E + APS D R G A +G EP P
Sbjct: 185 CHFIHNADE--RRPAPSGGGGASGDLRAFGARDALHLGFAREPRP 227
Score = 44.3 bits (103), Expect = 4e-04
Identities = 22/42 (52%), Positives = 27/42 (63%), Gaps = 2/42 (4%)
Query: 256 GGHGHH-GSNNFKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
GG G S +KT+LC F + GTC +GE+C FAHG ELR
Sbjct: 116 GGSGSQINSTRYKTELCRPFEESGTCKYGEKCQFAHGFHELR 157
Score = 40.4 bits (93), Expect = 0.005
Identities = 27/78 (34%), Positives = 34/78 (42%), Gaps = 10/78 (12%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWEL------GKHIAPSFDDHRPIGHAPAGR---- 150
KT +C F + CK+G+KC FAHG EL K+ IG P G
Sbjct: 128 KTELCRPFEESGTCKYGEKCQFAHGFHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHF 187
Query: 151 IGGRMEPPPGPATGFGAN 168
I E P P+ G GA+
Sbjct: 188 IHNADERRPAPSGGGGAS 205
Score = 35.0 bits (79), Expect = 0.22
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 7/45 (15%)
Query: 258 HGHHGSNN------FKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
HG H + +KT+LC F G C +G RCHF H E R
Sbjct: 151 HGFHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIHNADERR 195
>ROK_RAT (P61980) Heterogeneous nuclear ribonucleoprotein K
(dC-stretch binding protein) (CSBP)
Length = 463
Score = 49.3 bits (116), Expect = 1e-05
Identities = 42/162 (25%), Positives = 69/162 (41%), Gaps = 23/162 (14%)
Query: 83 RNVAAPPPPVPNGSTPAV---KTRICNKFNTAEGCKFGDKCHFA-----HGEWELGKHI- 133
RN+ PPPP P G + R ++++ G + A EW++
Sbjct: 305 RNLPLPPPPPPRGGDLMAYDRRGRPGDRYDGMVGFSADETWDSAIDTWSPSEWQMAYEPQ 364
Query: 134 -APSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSK 192
+D G G +GG + T ++++ LAG+IIGKGG K
Sbjct: 365 GGSGYDYSYAGGRGSYGDLGGPI-------------ITTQVTIPKDLAGSIIGKGGQRIK 411
Query: 193 QICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKD 234
QI ++GA + I E R I + G+ DQI+ A ++++
Sbjct: 412 QIRHESGASIKIDEPLEGSEDRIITITGTQDQIQNAQYLLQN 453
Score = 32.7 bits (73), Expect = 1.1
Identities = 21/82 (25%), Positives = 42/82 (50%), Gaps = 9/82 (10%)
Query: 172 KISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNM 231
+I +++ AGA+IGKGG N K + A +S+ + + +I + I+ +
Sbjct: 46 RILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSGPERILSISAD-----IETIGEI 100
Query: 232 VKDLLLT----LQMSAPPKSNQ 249
+K ++ T LQ+ +P ++Q
Sbjct: 101 LKKIIPTLEEGLQLPSPTATSQ 122
>ROK_RABIT (O19049) Heterogeneous nuclear ribonucleoprotein K (hnRNP
K)
Length = 463
Score = 49.3 bits (116), Expect = 1e-05
Identities = 42/162 (25%), Positives = 69/162 (41%), Gaps = 23/162 (14%)
Query: 83 RNVAAPPPPVPNGSTPAV---KTRICNKFNTAEGCKFGDKCHFA-----HGEWELGKHI- 133
RN+ PPPP P G + R ++++ G + A EW++
Sbjct: 305 RNLPLPPPPPPRGGDLMAYDRRGRPGDRYDGMVGFSADETWDSAIDTWSPSEWQMAYEPQ 364
Query: 134 -APSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSK 192
+D G G +GG + T ++++ LAG+IIGKGG K
Sbjct: 365 GGSGYDYSYAGGRGSYGDLGGPI-------------ITTQVTIPKDLAGSIIGKGGQRIK 411
Query: 193 QICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKD 234
QI ++GA + I E R I + G+ DQI+ A ++++
Sbjct: 412 QIRHESGASIKIDEPLEGSEDRIITITGTQDQIQNAQYLLQN 453
Score = 32.7 bits (73), Expect = 1.1
Identities = 21/82 (25%), Positives = 42/82 (50%), Gaps = 9/82 (10%)
Query: 172 KISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNM 231
+I +++ AGA+IGKGG N K + A +S+ + + +I + I+ +
Sbjct: 46 RILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSGPERILSISAD-----IETIGEI 100
Query: 232 VKDLLLT----LQMSAPPKSNQ 249
+K ++ T LQ+ +P ++Q
Sbjct: 101 LKKIIPTLEEGLQLPSPTATSQ 122
>ROK_MOUSE (P61979) Heterogeneous nuclear ribonucleoprotein K
Length = 463
Score = 49.3 bits (116), Expect = 1e-05
Identities = 42/162 (25%), Positives = 69/162 (41%), Gaps = 23/162 (14%)
Query: 83 RNVAAPPPPVPNGSTPAV---KTRICNKFNTAEGCKFGDKCHFA-----HGEWELGKHI- 133
RN+ PPPP P G + R ++++ G + A EW++
Sbjct: 305 RNLPLPPPPPPRGGDLMAYDRRGRPGDRYDGMVGFSADETWDSAIDTWSPSEWQMAYEPQ 364
Query: 134 -APSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSK 192
+D G G +GG + T ++++ LAG+IIGKGG K
Sbjct: 365 GGSGYDYSYAGGRGSYGDLGGPI-------------ITTQVTIPKDLAGSIIGKGGQRIK 411
Query: 193 QICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKD 234
QI ++GA + I E R I + G+ DQI+ A ++++
Sbjct: 412 QIRHESGASIKIDEPLEGSEDRIITITGTQDQIQNAQYLLQN 453
Score = 32.7 bits (73), Expect = 1.1
Identities = 21/82 (25%), Positives = 42/82 (50%), Gaps = 9/82 (10%)
Query: 172 KISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNM 231
+I +++ AGA+IGKGG N K + A +S+ + + +I + I+ +
Sbjct: 46 RILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSGPERILSISAD-----IETIGEI 100
Query: 232 VKDLLLT----LQMSAPPKSNQ 249
+K ++ T LQ+ +P ++Q
Sbjct: 101 LKKIIPTLEEGLQLPSPTATSQ 122
>ROK_HUMAN (P61978) Heterogeneous nuclear ribonucleoprotein K (hnRNP
K) (Transformation up-regulated nuclear protein) (TUNP)
Length = 463
Score = 49.3 bits (116), Expect = 1e-05
Identities = 42/162 (25%), Positives = 69/162 (41%), Gaps = 23/162 (14%)
Query: 83 RNVAAPPPPVPNGSTPAV---KTRICNKFNTAEGCKFGDKCHFA-----HGEWELGKHI- 133
RN+ PPPP P G + R ++++ G + A EW++
Sbjct: 305 RNLPLPPPPPPRGGDLMAYDRRGRPGDRYDGMVGFSADETWDSAIDTWSPSEWQMAYEPQ 364
Query: 134 -APSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSK 192
+D G G +GG + T ++++ LAG+IIGKGG K
Sbjct: 365 GGSGYDYSYAGGRGSYGDLGGPI-------------ITTQVTIPKDLAGSIIGKGGQRIK 411
Query: 193 QICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKD 234
QI ++GA + I E R I + G+ DQI+ A ++++
Sbjct: 412 QIRHESGASIKIDEPLEGSEDRIITITGTQDQIQNAQYLLQN 453
Score = 32.7 bits (73), Expect = 1.1
Identities = 21/82 (25%), Positives = 42/82 (50%), Gaps = 9/82 (10%)
Query: 172 KISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNM 231
+I +++ AGA+IGKGG N K + A +S+ + + +I + I+ +
Sbjct: 46 RILLQSKNAGAVIGKGGKNIKALRTDYNASVSVPDSSGPERILSISAD-----IETIGEI 100
Query: 232 VKDLLLT----LQMSAPPKSNQ 249
+K ++ T LQ+ +P ++Q
Sbjct: 101 LKKIIPTLEEGLQLPSPTATSQ 122
>TISD_HUMAN (P47974) Butyrate response factor 2 (TIS11D protein)
(EGF-response factor 2) (ERF-2)
Length = 492
Score = 48.9 bits (115), Expect = 1e-05
Identities = 43/163 (26%), Positives = 61/163 (37%), Gaps = 35/163 (21%)
Query: 3 FRKRGRPEPGFNSNGGIKKSKQELESLSSGVGS---KSKPCTKFFSTAGCPFGESCHFLH 59
FR R E G S + +Q+ S + S K++ C F + C +GE C F H
Sbjct: 119 FRDRSFSENGDRSQHLLHLQQQQKGGGGSQINSTRYKTELCRPFEESGTCKYGEKCQFAH 178
Query: 60 YVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDK 119
G++ + + P KT +C F+T C +G +
Sbjct: 179 ----GFHELRSLTR-----------------------HPKYKTELCRTFHTIGFCPYGPR 211
Query: 120 CHFAHGEWELGKHIAPS---FDDHRPIGHAPAGRIGGRMEPPP 159
CHF H E + APS D R G A +G EP P
Sbjct: 212 CHFIHNADE--RRPAPSGGASGDLRAFGTRDALHLGFPREPRP 252
Score = 45.4 bits (106), Expect = 2e-04
Identities = 21/41 (51%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query: 256 GGHGHHGSNNFKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
GG S +KT+LC F + GTC +GE+C FAHG ELR
Sbjct: 144 GGGSQINSTRYKTELCRPFEESGTCKYGEKCQFAHGFHELR 184
Score = 35.0 bits (79), Expect = 0.22
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 7/45 (15%)
Query: 258 HGHHGSNN------FKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
HG H + +KT+LC F G C +G RCHF H E R
Sbjct: 178 HGFHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIHNADERR 222
>FUB3_HUMAN (Q96I24) Far upstream element binding protein 3 (FUSE
binding protein 3)
Length = 572
Score = 48.9 bits (115), Expect = 1e-05
Identities = 29/93 (31%), Positives = 45/93 (48%), Gaps = 3/93 (3%)
Query: 174 SVEASLAGAIIGKGGVNSKQICRQTGAKLSIREH---ESDPNLRNIELEGSFDQIKEASN 230
+V A G +IGKGG N K I +Q+GA + ++ + SDPNLR + G QI+ A
Sbjct: 360 TVPADKCGLVIGKGGENIKSINQQSGAHVELQRNPPPNSDPNLRRFTIRGVPQQIEVARQ 419
Query: 231 MVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGS 263
++ + + + AP Q P H +
Sbjct: 420 LIDEKVGGTNLGAPGAFGQSPFSQPPAPPHQNT 452
Score = 37.7 bits (86), Expect = 0.034
Identities = 25/91 (27%), Positives = 47/91 (51%), Gaps = 3/91 (3%)
Query: 149 GRIGGRMEPPPGPATGFGANATAK-ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREH 207
G+I R PG +N+T + I + AS G +IG+GG KQ+ +TG K+ + +
Sbjct: 142 GQIVDRCRNGPGFHNDIDSNSTIQEILIPASKVGLVIGRGGETIKQLQERTGVKMVMIQD 201
Query: 208 ESDPN--LRNIELEGSFDQIKEASNMVKDLL 236
P + + + G ++++A MV +++
Sbjct: 202 GPLPTGADKPLRITGDAFKVQQAREMVLEII 232
Score = 36.2 bits (82), Expect = 0.10
Identities = 21/88 (23%), Positives = 41/88 (45%), Gaps = 4/88 (4%)
Query: 172 KISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNM 231
++SV G +IG+ G K+I G ++ + + R ++ G D+ + A+++
Sbjct: 257 EVSVPRFAVGIVIGRNGEMIKKIQNDAGVRIQFKPDDGISPERAAQVMGPPDRCQHAAHI 316
Query: 232 VKDLLLTLQMSAPPKSNQGGAGGPGGHG 259
+ +L+LT Q + GG G G
Sbjct: 317 ISELILTAQ----ERDGFGGLAAARGRG 340
Score = 33.9 bits (76), Expect = 0.50
Identities = 24/96 (25%), Positives = 41/96 (42%), Gaps = 12/96 (12%)
Query: 170 TAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEAS 229
T + V + G IIG+GG +I ++G K+ I S R L G+ + I++A
Sbjct: 79 TEEFKVPDKMVGFIIGRGGEQISRIQAESGCKIQIASESSGIPERPCVLTGTPESIEQAK 138
Query: 230 NMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNN 265
++ ++ + GPG H SN+
Sbjct: 139 RLLGQIVDRCR------------NGPGFHNDIDSNS 162
>TIS1_DROME (P47980) TIS11 protein (dTIS11)
Length = 437
Score = 47.4 bits (111), Expect = 4e-05
Identities = 21/47 (44%), Positives = 26/47 (54%)
Query: 83 RNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
R + P PP +T KT +C F A CK+G+KC FAHG EL
Sbjct: 120 RTQSEPLPPQQPMNTSRYKTELCRPFEEAGECKYGEKCQFAHGSHEL 166
Score = 40.8 bits (94), Expect = 0.004
Identities = 17/34 (50%), Positives = 24/34 (70%), Gaps = 1/34 (2%)
Query: 263 SNNFKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
++ +KT+LC F + G C +GE+C FAHG ELR
Sbjct: 134 TSRYKTELCRPFEEAGECKYGEKCQFAHGSHELR 167
Score = 38.1 bits (87), Expect = 0.026
Identities = 24/89 (26%), Positives = 32/89 (34%), Gaps = 27/89 (30%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNG 95
K++ C F C +GE C F H G + RNV
Sbjct: 138 KTELCRPFEEAGECKYGEKCQFAH---GSHEL---------------RNV---------H 170
Query: 96 STPAVKTRICNKFNTAEGCKFGDKCHFAH 124
P KT C F++ C +G +CHF H
Sbjct: 171 RHPKYKTEYCRTFHSVGFCPYGPRCHFVH 199
Score = 36.6 bits (83), Expect = 0.077
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 7/45 (15%)
Query: 258 HGHHGSNN------FKTKLCENF-AKGTCTFGERCHFAHGPAELR 295
HG H N +KT+ C F + G C +G RCHF H E R
Sbjct: 161 HGSHELRNVHRHPKYKTEYCRTFHSVGFCPYGPRCHFVHNADEAR 205
Score = 32.7 bits (73), Expect = 1.1
Identities = 13/35 (37%), Positives = 19/35 (54%)
Query: 25 ELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLH 59
EL ++ K++ C F S CP+G CHF+H
Sbjct: 165 ELRNVHRHPKYKTEYCRTFHSVGFCPYGPRCHFVH 199
>TTP_RAT (P47973) Tristetraproline (TTP) (Zinc finger protein 36)
(Zfp-36) (TIS11A protein) (TIS11)
Length = 320
Score = 44.3 bits (103), Expect = 4e-04
Identities = 17/35 (48%), Positives = 26/35 (73%), Gaps = 1/35 (2%)
Query: 263 SNNFKTKLCENFAK-GTCTFGERCHFAHGPAELRK 296
S+ +KT+LC +++ G C +G +C FAHGP ELR+
Sbjct: 94 SSRYKTELCRTYSESGRCRYGAKCQFAHGPGELRQ 128
Score = 43.5 bits (101), Expect = 6e-04
Identities = 35/120 (29%), Positives = 52/120 (43%), Gaps = 18/120 (15%)
Query: 12 GFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHM 71
G S+GG+ +S+ SGV S+ + G SC ++ PG
Sbjct: 23 GTESSGGLWNINSS-DSIPSGVTSRLTG-----RSTSLVEGRSCSWVPPPPG-------- 68
Query: 72 MNLAPSAQAP-PRNVAAPPPPVPNGSTPA-VKTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
AP A P P +P P +T + KT +C ++ + C++G KC FAHG EL
Sbjct: 69 --FAPLAPRPGPELSPSPTSPTATPTTSSRYKTELCRTYSESGRCRYGAKCQFAHGPGEL 126
Score = 39.3 bits (90), Expect = 0.012
Identities = 15/29 (51%), Positives = 19/29 (64%), Gaps = 1/29 (3%)
Query: 266 FKTKLCENF-AKGTCTFGERCHFAHGPAE 293
+KT+LC F +G C +G RCHF H P E
Sbjct: 135 YKTELCHKFYLQGRCPYGSRCHFIHNPTE 163
Score = 37.7 bits (86), Expect = 0.034
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 11/73 (15%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLH------YVPGGYNAVAHMMNLAPSAQAPPRNVAAPP 89
K++ C KF+ CP+G CHF+H +PG H++ + S P P
Sbjct: 136 KTELCHKFYLQGRCPYGSRCHFIHNPTEDLALPG----QPHVLRQSISFSGLPSGRRTSP 191
Query: 90 PPVPNGSTPAVKT 102
PP P S P++ +
Sbjct: 192 PP-PGFSGPSLSS 203
Score = 37.0 bits (84), Expect = 0.059
Identities = 22/93 (23%), Positives = 31/93 (32%), Gaps = 27/93 (29%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNG 95
K++ C + + C +G C F H A H
Sbjct: 98 KTELCRTYSESGRCRYGAKCQFAHGPGELRQANRH------------------------- 132
Query: 96 STPAVKTRICNKFNTAEGCKFGDKCHFAHGEWE 128
P KT +C+KF C +G +CHF H E
Sbjct: 133 --PKYKTELCHKFYLQGRCPYGSRCHFIHNPTE 163
>TTP_HUMAN (P26651) Tristetraproline (TTP) (Zinc finger protein 36
homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth
factor-inducible nuclear protein NUP475) (G0/G1 switch
regulatory protein 24)
Length = 326
Score = 44.3 bits (103), Expect = 4e-04
Identities = 37/120 (30%), Positives = 53/120 (43%), Gaps = 17/120 (14%)
Query: 11 PGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAH 70
PG+ S+G S + S SGV S+ + G SC ++ PG
Sbjct: 30 PGWGSSGPWSLSPSD--SSPSGVTSRLPG-----RSTSLVEGRSCGWVPPPPG------- 75
Query: 71 MMNLAPSAQAPPRNVAAPPPPVPNGSTPA-VKTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
LAP + P +P P +TP+ KT +C F+ + C++G KC FAHG EL
Sbjct: 76 FAPLAP--RLGPELSPSPTSPTATSTTPSRYKTELCRTFSESGRCRYGAKCQFAHGLGEL 133
Score = 43.9 bits (102), Expect = 5e-04
Identities = 37/151 (24%), Positives = 52/151 (33%), Gaps = 45/151 (29%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNG 95
K++ C F + C +G C F H + A H
Sbjct: 105 KTELCRTFSESGRCRYGAKCQFAHGLGELRQANRH------------------------- 139
Query: 96 STPAVKTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPIGHAP-------- 147
P KT +C+KF C +G +CHF H PS D P GH P
Sbjct: 140 --PKYKTELCHKFYLQGRCPYGSRCHFIHN---------PSEDLAAP-GHPPVLRQSISF 187
Query: 148 AGRIGGRMEPPPGPATGFGANATAKISVEAS 178
+G GR PP P + +++ S +S
Sbjct: 188 SGLPSGRRTSPPPPGLAGPSLSSSSFSPSSS 218
Score = 40.0 bits (92), Expect = 0.007
Identities = 16/34 (47%), Positives = 24/34 (70%), Gaps = 1/34 (2%)
Query: 264 NNFKTKLCENFAK-GTCTFGERCHFAHGPAELRK 296
+ +KT+LC F++ G C +G +C FAHG ELR+
Sbjct: 102 SRYKTELCRTFSESGRCRYGAKCQFAHGLGELRQ 135
Score = 39.7 bits (91), Expect = 0.009
Identities = 15/29 (51%), Positives = 20/29 (68%), Gaps = 1/29 (3%)
Query: 266 FKTKLCENF-AKGTCTFGERCHFAHGPAE 293
+KT+LC F +G C +G RCHF H P+E
Sbjct: 142 YKTELCHKFYLQGRCPYGSRCHFIHNPSE 170
>TISB_HUMAN (Q07352) Butyrate response factor 1 (TIS11B protein)
(EGF-response factor 1) (ERF-1)
Length = 338
Score = 44.3 bits (103), Expect = 4e-04
Identities = 21/42 (50%), Positives = 28/42 (66%), Gaps = 2/42 (4%)
Query: 255 PGGHGHHGSNNFKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
PGG G S+ +KT+LC F + G C +G++C FAHG ELR
Sbjct: 105 PGG-GQVNSSRYKTELCRPFEENGACKYGDKCQFAHGIHELR 145
Score = 40.8 bits (94), Expect = 0.004
Identities = 22/93 (23%), Positives = 32/93 (33%), Gaps = 27/93 (29%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNG 95
K++ C F C +G+ C F H + + H
Sbjct: 116 KTELCRPFEENGACKYGDKCQFAHGIHELRSLTRH------------------------- 150
Query: 96 STPAVKTRICNKFNTAEGCKFGDKCHFAHGEWE 128
P KT +C F+T C +G +CHF H E
Sbjct: 151 --PKYKTELCRTFHTIGFCPYGPRCHFIHNAEE 181
Score = 40.0 bits (92), Expect = 0.007
Identities = 16/29 (55%), Positives = 18/29 (61%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
KT +C F CK+GDKC FAHG EL
Sbjct: 116 KTELCRPFEENGACKYGDKCQFAHGIHEL 144
Score = 35.0 bits (79), Expect = 0.22
Identities = 18/55 (32%), Positives = 25/55 (44%), Gaps = 8/55 (14%)
Query: 13 FNSNGGIKKSKQ--------ELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLH 59
F NG K + EL SL+ K++ C F + CP+G CHF+H
Sbjct: 123 FEENGACKYGDKCQFAHGIHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIH 177
Score = 35.0 bits (79), Expect = 0.22
Identities = 15/31 (48%), Positives = 18/31 (57%), Gaps = 1/31 (3%)
Query: 266 FKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
+KT+LC F G C +G RCHF H E R
Sbjct: 153 YKTELCRTFHTIGFCPYGPRCHFIHNAEERR 183
>FUB2_HUMAN (Q92945) Far upstream element binding protein 2 (FUSE
binding protein 2) (KH type splicing regulatory protein)
(KSRP) (p75)
Length = 707
Score = 43.9 bits (102), Expect = 5e-04
Identities = 29/112 (25%), Positives = 49/112 (42%), Gaps = 18/112 (16%)
Query: 153 GRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPN 212
G + PPP + T + V + G IIG+GG +I + +G K+ I
Sbjct: 135 GPIHPPPR------TSMTEEYRVPDGMVGLIIGRGGEQINKIQQDSGCKVQISPDSGGLP 188
Query: 213 LRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSN 264
R++ L G+ + +++A M+ D++ G GGP G H +N
Sbjct: 189 ERSVSLTGAPESVQKAKMMLDDIV------------SRGRGGPPGQFHDNAN 228
Score = 42.7 bits (99), Expect = 0.001
Identities = 27/95 (28%), Positives = 46/95 (48%), Gaps = 3/95 (3%)
Query: 173 ISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMV 232
+ V G +IG+ G K+I G ++ ++ + + + G D+ + A+ ++
Sbjct: 327 VPVPRHSVGVVIGRSGEMIKKIQNDAGVRIQFKQDDGTGPEKIAHIMGPPDRCEHAARII 386
Query: 233 KDLLLTLQMSAPPKSNQGGAGGPGGHGH-HGSNNF 266
DLL +L+ S PP GG G PGG G G N+
Sbjct: 387 NDLLQSLR-SGPP-GPPGGPGIPGGRGRGRGQGNW 419
Score = 41.6 bits (96), Expect = 0.002
Identities = 37/122 (30%), Positives = 51/122 (41%), Gaps = 14/122 (11%)
Query: 147 PAGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIRE 206
P GR GR + GP G T S+ G +IG+GG N K I +QTGA + I
Sbjct: 407 PGGRGRGRGQGNWGP----GGEMT--FSIPTHKCGLVIGRGGENVKAINQQTGAFVEISR 460
Query: 207 H---ESDPNLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGS 263
P + + GS QI ++++ ++ P G GGPG G G
Sbjct: 461 QLPPTGTPTSKLFIIRGSPQQIDHCRQLIEE-----KIEGPLCPVGPGPGGPGPAGPMGP 515
Query: 264 NN 265
N
Sbjct: 516 FN 517
Score = 40.8 bits (94), Expect = 0.004
Identities = 30/106 (28%), Positives = 52/106 (48%), Gaps = 6/106 (5%)
Query: 158 PPGP----ATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAK-LSIREHESDPN 212
PPG A G +I + A AG +IGKGG KQ+ + G K + I++ + N
Sbjct: 219 PPGQFHDNANGGQNGTVQEIMIPAGKAGLVIGKGGETIKQLQERAGVKMILIQDGSQNTN 278
Query: 213 L-RNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGG 257
+ + + + G ++++A MV D+L + + N+ G+ GG
Sbjct: 279 VDKPLRIIGDPYKVQQACEMVMDILRNVTKAGFGDRNEYGSRIGGG 324
>TISB_RAT (P17431) Butyrate response factor 1 (TIS11B protein)
(EGF-inducible protein CMG1)
Length = 338
Score = 43.5 bits (101), Expect = 6e-04
Identities = 19/40 (47%), Positives = 26/40 (64%), Gaps = 1/40 (2%)
Query: 257 GHGHHGSNNFKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
G G S+ +KT+LC F + G C +G++C FAHG ELR
Sbjct: 106 GSGQVNSSRYKTELCRPFEENGACKYGDKCQFAHGIHELR 145
Score = 41.6 bits (96), Expect = 0.002
Identities = 28/117 (23%), Positives = 41/117 (34%), Gaps = 30/117 (25%)
Query: 15 SNGGIKKSKQELESLSSGVGS---KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHM 71
S GG + + + S V S K++ C F C +G+ C F H + + H
Sbjct: 92 SEGGERLLPTQKQPGSGQVNSSRYKTELCRPFEENGACKYGDKCQFAHGIHELRSLTRH- 150
Query: 72 MNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAHGEWE 128
P KT +C F+T C +G +CHF H E
Sbjct: 151 --------------------------PKYKTELCRTFHTIGFCPYGPRCHFIHNAEE 181
Score = 40.0 bits (92), Expect = 0.007
Identities = 16/29 (55%), Positives = 18/29 (61%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
KT +C F CK+GDKC FAHG EL
Sbjct: 116 KTELCRPFEENGACKYGDKCQFAHGIHEL 144
Score = 35.0 bits (79), Expect = 0.22
Identities = 18/55 (32%), Positives = 25/55 (44%), Gaps = 8/55 (14%)
Query: 13 FNSNGGIKKSKQ--------ELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLH 59
F NG K + EL SL+ K++ C F + CP+G CHF+H
Sbjct: 123 FEENGACKYGDKCQFAHGIHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIH 177
Score = 35.0 bits (79), Expect = 0.22
Identities = 15/31 (48%), Positives = 18/31 (57%), Gaps = 1/31 (3%)
Query: 266 FKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
+KT+LC F G C +G RCHF H E R
Sbjct: 153 YKTELCRTFHTIGFCPYGPRCHFIHNAEERR 183
>TISB_MOUSE (P23950) Butyrate response factor 1 (TIS11B protein)
Length = 338
Score = 43.5 bits (101), Expect = 6e-04
Identities = 19/40 (47%), Positives = 26/40 (64%), Gaps = 1/40 (2%)
Query: 257 GHGHHGSNNFKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
G G S+ +KT+LC F + G C +G++C FAHG ELR
Sbjct: 106 GSGQVNSSRYKTELCRPFEENGACKYGDKCQFAHGIHELR 145
Score = 41.6 bits (96), Expect = 0.002
Identities = 28/117 (23%), Positives = 41/117 (34%), Gaps = 30/117 (25%)
Query: 15 SNGGIKKSKQELESLSSGVGS---KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHM 71
S GG + + + S V S K++ C F C +G+ C F H + + H
Sbjct: 92 SEGGERLLPTQKQPGSGQVNSSRYKTELCRPFEENGACKYGDKCQFAHGIHELRSLTRH- 150
Query: 72 MNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAHGEWE 128
P KT +C F+T C +G +CHF H E
Sbjct: 151 --------------------------PKYKTELCRTFHTIGFCPYGPRCHFIHNAEE 181
Score = 40.0 bits (92), Expect = 0.007
Identities = 16/29 (55%), Positives = 18/29 (61%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
KT +C F CK+GDKC FAHG EL
Sbjct: 116 KTELCRPFEENGACKYGDKCQFAHGIHEL 144
Score = 35.0 bits (79), Expect = 0.22
Identities = 18/55 (32%), Positives = 25/55 (44%), Gaps = 8/55 (14%)
Query: 13 FNSNGGIKKSKQ--------ELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLH 59
F NG K + EL SL+ K++ C F + CP+G CHF+H
Sbjct: 123 FEENGACKYGDKCQFAHGIHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIH 177
Score = 35.0 bits (79), Expect = 0.22
Identities = 15/31 (48%), Positives = 18/31 (57%), Gaps = 1/31 (3%)
Query: 266 FKTKLCENFAK-GTCTFGERCHFAHGPAELR 295
+KT+LC F G C +G RCHF H E R
Sbjct: 153 YKTELCRTFHTIGFCPYGPRCHFIHNAEERR 183
>CTH2_YEAST (P47977) Zinc finger protein CTH2 (YTIS11 protein)
Length = 285
Score = 42.7 bits (99), Expect = 0.001
Identities = 17/31 (54%), Positives = 24/31 (76%), Gaps = 1/31 (3%)
Query: 266 FKTKLCENFA-KGTCTFGERCHFAHGPAELR 295
+KT+LCE+F KG+C +G +C FAHG EL+
Sbjct: 170 YKTELCESFTLKGSCPYGSKCQFAHGLGELK 200
Score = 40.8 bits (94), Expect = 0.004
Identities = 25/82 (30%), Positives = 32/82 (38%), Gaps = 12/82 (14%)
Query: 60 YVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVK------------TRICNK 107
YVP NL +Q P+N A+ + + P VK T +C
Sbjct: 118 YVPKSSQLPLTSQNLQQLSQQKPKNDASFSSEKESSAQPKVKSQVQETPKQLYKTELCES 177
Query: 108 FNTAEGCKFGDKCHFAHGEWEL 129
F C +G KC FAHG EL
Sbjct: 178 FTLKGSCPYGSKCQFAHGLGEL 199
Score = 34.7 bits (78), Expect = 0.29
Identities = 15/27 (55%), Positives = 18/27 (66%), Gaps = 1/27 (3%)
Query: 265 NFKTKLCENFAK-GTCTFGERCHFAHG 290
NF+TK C N+ K G C +G RC F HG
Sbjct: 207 NFRTKPCVNWEKLGYCPYGRRCCFKHG 233
>TTP_SHEEP (Q6S9E0) Tristetraproline (TTP) (Zinc finger protein 36
homolog) (Zfp-36)
Length = 325
Score = 42.4 bits (98), Expect = 0.001
Identities = 27/82 (32%), Positives = 37/82 (44%), Gaps = 16/82 (19%)
Query: 52 GESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAV----KTRICNK 107
G SC ++ PG AP A P ++ P P +TP KT +C
Sbjct: 62 GRSCGWVPPPPG----------FAPLAPRPSSELS--PSPTSPTATPTTSSRYKTELCRT 109
Query: 108 FNTAEGCKFGDKCHFAHGEWEL 129
F+ + C++G KC FAHG EL
Sbjct: 110 FSESGRCRYGAKCQFAHGLGEL 131
Score = 41.6 bits (96), Expect = 0.002
Identities = 17/35 (48%), Positives = 25/35 (70%), Gaps = 1/35 (2%)
Query: 263 SNNFKTKLCENFAK-GTCTFGERCHFAHGPAELRK 296
S+ +KT+LC F++ G C +G +C FAHG ELR+
Sbjct: 99 SSRYKTELCRTFSESGRCRYGAKCQFAHGLGELRQ 133
Score = 40.8 bits (94), Expect = 0.004
Identities = 36/141 (25%), Positives = 48/141 (33%), Gaps = 52/141 (36%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNG 95
K++ C F + C +G C F H + + P R+
Sbjct: 103 KTELCRTFSESGRCRYGAKCQFAHGLG--------------ELRQPSRH----------- 137
Query: 96 STPAVKTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPIGH---------- 145
P KT +C+KF C +G +CHF H PS D P GH
Sbjct: 138 --PKYKTELCHKFYLQGRCPYGSRCHFIHN---------PSEDLAAP-GHPHVLRQSISF 185
Query: 146 --APAGRIGGRMEPPPGPATG 164
P+GR R PPP G
Sbjct: 186 SGLPSGR---RTSPPPASLAG 203
Score = 39.7 bits (91), Expect = 0.009
Identities = 15/29 (51%), Positives = 20/29 (68%), Gaps = 1/29 (3%)
Query: 266 FKTKLCENF-AKGTCTFGERCHFAHGPAE 293
+KT+LC F +G C +G RCHF H P+E
Sbjct: 140 YKTELCHKFYLQGRCPYGSRCHFIHNPSE 168
Score = 37.7 bits (86), Expect = 0.034
Identities = 27/96 (28%), Positives = 40/96 (41%), Gaps = 22/96 (22%)
Query: 25 ELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLH------YVPGGYNAVAHMMNLA--P 76
EL S K++ C KF+ CP+G CHF+H PG + + ++ + P
Sbjct: 130 ELRQPSRHPKYKTELCHKFYLQGRCPYGSRCHFIHNPSEDLAAPGHPHVLRQSISFSGLP 189
Query: 77 SAQ---APPRNVAAP-----------PPPVPNGSTP 98
S + PP ++A P PP P G P
Sbjct: 190 SGRRTSPPPASLAGPSVPSWSFSPSSSPPPPPGDLP 225
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.135 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,455,213
Number of Sequences: 164201
Number of extensions: 2052483
Number of successful extensions: 10987
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 60
Number of HSP's that attempted gapping in prelim test: 10561
Number of HSP's gapped (non-prelim): 452
length of query: 296
length of database: 59,974,054
effective HSP length: 109
effective length of query: 187
effective length of database: 42,076,145
effective search space: 7868239115
effective search space used: 7868239115
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0221.10


