

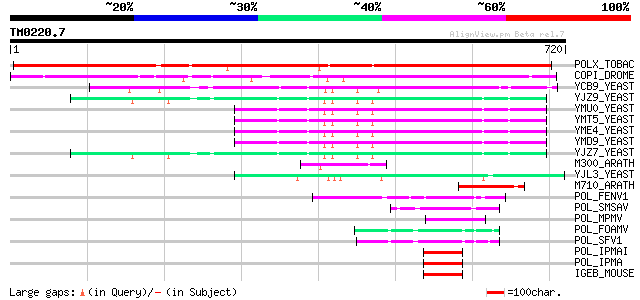
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0220.7
(720 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 579 e-164
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 287 8e-77
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 132 2e-30
YJZ9_YEAST (P47100) Transposon Ty1 protein B 112 3e-24
YMU0_YEAST (Q04670) Transposon Ty1 protein B 112 3e-24
YMT5_YEAST (Q04214) Transposon Ty1 protein B 112 3e-24
YME4_YEAST (Q04711) Transposon Ty1 protein B 112 3e-24
YMD9_YEAST (Q03434) Transposon Ty1 protein B 112 3e-24
YJZ7_YEAST (P47098) Transposon Ty1 protein B 111 8e-24
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 80 2e-14
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 79 3e-14
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 79 5e-14
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 49 6e-05
POL_SMSAV (P03359) Pol polyprotein [Contains: Reverse transcript... 47 1e-04
POL_MPMV (P07572) Pol polyprotein [Contains: Reverse transcripta... 47 2e-04
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 47 2e-04
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 47 2e-04
POL_IPMAI (P12894) Probable Pol polyprotein [Contains: Integrase... 47 2e-04
POL_IPMA (P11368) Putative Pol polyprotein [Contains: Integrase ... 47 2e-04
IGEB_MOUSE (P03975) IgE-binding protein 47 2e-04
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 579 bits (1492), Expect = e-164
Identities = 305/714 (42%), Positives = 446/714 (61%), Gaps = 27/714 (3%)
Query: 5 KVKIERFDGRD-FGFWKMLMEDYLYQKMLYQPLT--GKKPNDMKQEDWDLLDRQALGVIR 61
K ++ +F+G + F W+ M D L Q+ L++ L KKP+ MK EDW LD +A IR
Sbjct: 5 KYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASAIR 64
Query: 62 LTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHLIRRLFNLRMGEGNSVTEHINS 121
L LS +V NI++E T + L ++Y NK++L ++L+ L M EG + H+N
Sbjct: 65 LHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHLNV 124
Query: 122 FNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSARDNKLKFDDIRDLILSE 181
FN +I+QL+++ + + E + LL SLP S+ T + + +LK D L+L+E
Sbjct: 125 FNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKTTIELK-DVTSALLLNE 183
Query: 182 DIRRKDSGESSNTFGSALNTESRGRGSQKS-HNQSQGRGRSKSRGRSQTRVRNDITCWNC 240
+R+K + G AL TE RGR Q+S +N + R KS+ RS++RVRN C+NC
Sbjct: 184 KMRKKPENQ-----GQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRN---CYNC 235
Query: 241 DRKGHFTNQCKAPRKKKNYQKR*DDDESANAATEEVADTLI--------CSLDSPVDSWV 292
++ GHF C PRK K +D++ A + + ++ L P WV
Sbjct: 236 NQPGHFKRDCPNPRKGKGETSGQKNDDNTAAMVQNNDNVVLFINEEEECMHLSGPESEWV 295
Query: 293 IDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRH 352
+D+ AS H P ++L Y+ G FG V + + I GIGDI I+++ G L +VRH
Sbjct: 296 VDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVLKDVRH 355
Query: 353 VPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYM----VAEEDMIA 408
VP ++ NLIS LD +GY + F W++TKG+LV+A+G RG+LY + + ++ A
Sbjct: 356 VPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGVARGTLYRTNAEICQGELNA 415
Query: 409 VTEAINSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCEHCILGKQRKVSFSKAG 468
+ I S +WH+R+GHMSEKG++I+A K +S K + C++C+ GKQ +VSF +
Sbjct: 416 AQDEI-SVDLWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCLFGKQHRVSFQTSS 474
Query: 469 RKSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWK 528
+ K L+LV++DV GP ++S+GG++Y+VTFIDD++RK+WVY LK+K VF VF+K+
Sbjct: 475 ER-KLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQKFH 533
Query: 529 TEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLN 588
VE +TG K+K L+SDNGGEY S+EF+++CS +GIR KT+PGTP+ NGVAERMNRT+
Sbjct: 534 ALVERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIV 593
Query: 589 ERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGC 648
E+ R M + LPK FW +A+ TA YLINR PSVPL +++PE VW KEVS SHLKVFGC
Sbjct: 594 EKVRSMLRMAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGC 653
Query: 649 VSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSINVTFNES 702
++ + ++R KLD K+I C FIGYG + +GYR WD +K+IRS +V F ES
Sbjct: 654 RAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRES 707
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 287 bits (734), Expect = 8e-77
Identities = 209/736 (28%), Positives = 365/736 (49%), Gaps = 50/736 (6%)
Query: 1 LEEGKVKIERFDGRDFGFWKMLMEDYLYQKMLYQPLTGKKPNDMKQEDWDLLDRQALGVI 60
+++ K I+ FDG + WK + L ++ + + + G PN++ + W +R A I
Sbjct: 1 MDKAKRNIKPFDGEKYAIWKFRIRALLAEQDVLKVVDGLMPNEV-DDSWKKAERCAKSTI 59
Query: 61 RLTLSKNVAFNIVNEKTTADLMKALSNMYEKPFAANKVHLIRRLFNLRMGEGNSVTEHIN 120
LS + ++ T +++ L +YE+ A+++ L +RL +L++ S+ H +
Sbjct: 60 IEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFH 119
Query: 121 SFNTIISQLSSVKITFDNELMVLSLLQSLPDSWAATVTAVSNSARDNKLKFDDIRDLILS 180
F+ +IS+L + + + LL +LP + +TA+ + +N L +++ +L
Sbjct: 120 IFDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEEN-LTLAFVKNRLLD 178
Query: 181 EDIRRKDSGESSNTFGSALNTESRGRGSQKSHNQSQGRGRSKSR---GRSQTRVRNDITC 237
++I+ K+ + ++T +N + +N + R + G S+ +V+ C
Sbjct: 179 QEIKIKN--DHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGNSKYKVK----C 232
Query: 238 WNCDRKGHFTNQCKAPRKKKNYQKR*DDDESANAATEEVADTLICSLD--SPVDS--WVI 293
+C R+GH C ++ N + + ++++ AT ++ ++ S +D+ +V+
Sbjct: 233 HHCGREGHIKKDCFHYKRILNNKNK-ENEKQVQTATSHGIAFMVKEVNNTSVMDNCGFVL 291
Query: 294 DSGASFHTIPSKELLSNYI---------CGKFGKVYLADGKPLDIVGIGDIDIRSSNGTL 344
DSGAS H I + L ++ + K G+ A + + +R N
Sbjct: 292 DSGASDHLINDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGI---------VRLRNDHE 342
Query: 345 WTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEE 404
TL +V NL+S+ +L + G F ++K L+V K G L V
Sbjct: 343 ITLEDVLFCKEAAGNLMSVKRLQEAGMSIEFDKSGVTISKNGLMVV--KNSGMLNNVPVI 400
Query: 405 DMIAVT---EAINSSSIWHQRLGHMSEKGM-----KIMASKGKMSNLKHVDLGVCEHCIL 456
+ A + + N+ +WH+R GH+S+ + K M S + N + +CE C+
Sbjct: 401 NFQAYSINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLN 460
Query: 457 GKQRKVSFSKAGRKSKSEK-LELVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKVWVYFLK 515
GKQ ++ F + K+ ++ L +VH+DV GP +L Y+V F+D T Y +K
Sbjct: 461 GKQARLPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIK 520
Query: 516 SKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPE 575
KSDVFS+F+ + + E LK+ L DNG EY S E ++FC + GI T+P TP+
Sbjct: 521 YKSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPHTPQ 580
Query: 576 QNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPL--DYQLPEEVW 633
NGV+ERM RT+ E+AR M + L K FW +A+ TA YLINR PS L + P E+W
Sbjct: 581 LNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRIPSRALVDSSKTPYEMW 640
Query: 634 YGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQNRKIIR 693
+ K+ L HL+VFG YV I +K+ K D K+ K F+GY + G++ WD N K I
Sbjct: 641 HNKKPYLKHLRVFGATVYVHI-KNKQGKFDDKSFKSIFVGY--EPNGFKLWDAVNEKFIV 697
Query: 694 SINVTFNESVLYKDRS 709
+ +V +E+ + R+
Sbjct: 698 ARDVVVDETNMVNSRA 713
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 132 bits (333), Expect = 2e-30
Identities = 148/648 (22%), Positives = 265/648 (40%), Gaps = 66/648 (10%)
Query: 104 LFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDNELMVLSLLQSLPDSW--------AA 155
L NL S + +TII +L I + L +L+ L +
Sbjct: 268 LANLEYDGSTSADTFEITVSTIIQRLKENNINVSDRLACQLILKGLSGDFKYLRNQYRTK 327
Query: 156 TVTAVSNSARDNKLKFDDIRDLILSEDIRRKDSGESSN---TFGSALNTESRGRGSQKSH 212
T +S + +L +D+ + + L++ + K E N T + NT+ R Q+++
Sbjct: 328 TNMKLSQLFAEIQLIYDENKIMNLNKPSQYKQHSEYKNVSRTSPNTTNTKVTTRNYQRTN 387
Query: 213 NQSQGRGRSKSRGRSQ--TRVRNDITCWNCDRKGHFTNQCKAPRKKKNYQKR*DDDESAN 270
+ ++ + S +RV ND H + + + D++ S
Sbjct: 388 SSKPRAAKAHNIATSSKFSRVNND----------HINESTVSSQYLSD-----DNELSLG 432
Query: 271 AATEEVADT-LICSLDSPVDSWVIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDI 329
+E T I S D D +IDSGAS + S L + + A + + I
Sbjct: 433 QQQKESKPTHTIDSNDELPDHLLIDSGASQTLVRSAHYLHHATPNSEINIVDAQKQDIPI 492
Query: 330 VGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVV 389
IG++ NGT ++ + H P I +L+S+ +L ++ F + + G V+
Sbjct: 493 NAIGNLHFNFQNGTKTSIKAL-HTPNIAYDLLSLSELANQNITACFTRNTLERSDGT-VL 550
Query: 390 ARGKKRGSLYMVAEEDMIA------VTEAINSSS--------IWHQRLGHMSEKGMKIMA 435
A K G Y ++++ +I +N S + H+ LGH + + ++
Sbjct: 551 APIVKHGDFYWLSKKYLIPSHISKLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSL 610
Query: 436 SKGKMSNLKHVDLG-------VCEHCILGKQRKVSFSKAGRKSKSEKLE---LVHTDVWG 485
K ++ LK D+ C C++GK K K R E E +HTD++G
Sbjct: 611 KKNAVTYLKESDIEWSNASTYQCPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFG 670
Query: 486 PAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSD--VFSVFKKWKTEVENQTGLKIKSLK 543
P Y+++F D+ TR WVY L + + + +VF ++NQ ++ ++
Sbjct: 671 PVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQ 730
Query: 544 SDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKM 603
D G EY ++ KF + GI T +GVAER+NRTL R + SGLP
Sbjct: 731 MDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLLNDCRTLLHCSGLPNH 790
Query: 604 FWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRD-KL 662
W A+ + + N S D + + ++ + FG V++++ D K+
Sbjct: 791 LWFSAVEFSTIIRNSLVSPKNDKSARQHAGLA-GLDITTILPFG--QPVIVNNHNPDSKI 847
Query: 663 DPKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSINVTFNESVLYKDRSS 710
P+ I + + + YGY + +K + + N V+ +D+ S
Sbjct: 848 HPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNY-----VILQDKQS 890
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 112 bits (281), Expect = 3e-24
Identities = 140/655 (21%), Positives = 250/655 (37%), Gaps = 54/655 (8%)
Query: 80 DLMKALSNMYEKPFA-ANKVHLIRRLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDN 138
D+MK LS EK + + + I L NL+ II +L++ I +N
Sbjct: 247 DIMKILSKSIEKMQSDTQEANDIVTLANLQYNGSTPADAFETKVTNIIDRLNNNGIHINN 306
Query: 139 ELMVLSLLQSLPDSWAATV--------TAVSNSARDNKLKFDDIRDLILSEDIRRKDSGE 190
++ +++ L + V+ D +++ + S+ R++ +
Sbjct: 307 KVACQLIMRGLSGEYKFLRYTRHRHLNMTVAELFLDIHAIYEEQQGSRNSKPNYRRNPSD 366
Query: 191 SSNTFGSALNTESR---GRGSQKSHNQSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFT 247
N S NT R QK++N R+ + S D +
Sbjct: 367 EKNDSRSYTNTTKPKVIARNPQKTNNSKSKTARAHNVSTSNNSPSTD--------NDSIS 418
Query: 248 NQCKAPRKKKNYQKR*DDDESANAATEEVADTLICSLDSPVDSWVIDSGASFHTIPSKEL 307
P + N D TE + S D ++DSGAS I S
Sbjct: 419 KSTTEPIQLNNKH----DLILGQKLTESTVNHTNHSDDELPGHLLLDSGASRTLIRSAHH 474
Query: 308 LSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLD 367
+ + V A + + I IGD+ + T ++ V H P I +L+S+ +L
Sbjct: 475 IHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI-KVLHTPNIAYDLLSLNELA 533
Query: 368 DEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMI-------AVTEAINSSS--- 417
F + + G V+A K G Y V+++ ++ + S S
Sbjct: 534 AVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNISVPTINNVHTSESTRK 592
Query: 418 ----IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG-------VCEHCILGKQRKVSFSK 466
H+ L H + + ++ ++ D+ C C++GK K K
Sbjct: 593 YPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIK 652
Query: 467 AGR---KSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSD--VF 521
R ++ E + +HTD++GP Y+++F D++T+ WVY L + + +
Sbjct: 653 GSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSIL 712
Query: 522 SVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAE 581
VF ++NQ + ++ D G EY ++ KF +NGI T +GVAE
Sbjct: 713 DVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAE 772
Query: 582 RMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLS 641
R+NRTL + R SGLP W AI + ++ + P + + + +S
Sbjct: 773 RLNRTLLDDCRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDIS 831
Query: 642 HLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSIN 696
L FG ++ D + K+ P+ I + + + YGY + +K + + N
Sbjct: 832 TLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTN 885
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 112 bits (280), Expect = 3e-24
Identities = 101/431 (23%), Positives = 180/431 (41%), Gaps = 30/431 (6%)
Query: 292 VIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVR 351
++DSGAS I S + + V A + + I IGD+ + T ++ V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI-KVL 90
Query: 352 HVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMI---- 407
H P I +L+S+ +L F + + G V+A K G Y V+++ ++
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 408 ---AVTEAINSSS-------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG-------V 450
+ S S H+ L H + + ++ ++ D+
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 209
Query: 451 CEHCILGKQRKVSFSKAGR---KSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTR 507
C C++GK K K R ++ E + +HTD++GP Y+++F D++T+
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 508 KVWVYFLKSKSD--VFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIR 565
WVY L + + + VF ++NQ + ++ D G EY ++ KF +NGI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 566 MIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLD 625
T +GVAER+NRTL + R SGLP W AI + ++ + P
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKS 388
Query: 626 YQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWD 685
+ + + +S L FG ++ D + K+ P+ I + + + YGY +
Sbjct: 389 KKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYL 447
Query: 686 EQNRKIIRSIN 696
+K + + N
Sbjct: 448 PSLKKTVDTTN 458
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 112 bits (280), Expect = 3e-24
Identities = 101/431 (23%), Positives = 180/431 (41%), Gaps = 30/431 (6%)
Query: 292 VIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVR 351
++DSGAS I S + + V A + + I IGD+ + T ++ V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI-KVL 90
Query: 352 HVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMI---- 407
H P I +L+S+ +L F + + G V+A K G Y V+++ ++
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 408 ---AVTEAINSSS-------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG-------V 450
+ S S H+ L H + + ++ ++ D+
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 209
Query: 451 CEHCILGKQRKVSFSKAGR---KSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTR 507
C C++GK K K R ++ E + +HTD++GP Y+++F D++T+
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 508 KVWVYFLKSKSD--VFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIR 565
WVY L + + + VF ++NQ + ++ D G EY ++ KF +NGI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 566 MIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLD 625
T +GVAER+NRTL + R SGLP W AI + ++ + P
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKS 388
Query: 626 YQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWD 685
+ + + +S L FG ++ D + K+ P+ I + + + YGY +
Sbjct: 389 KKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYL 447
Query: 686 EQNRKIIRSIN 696
+K + + N
Sbjct: 448 PSLKKTVDTTN 458
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 112 bits (280), Expect = 3e-24
Identities = 101/431 (23%), Positives = 180/431 (41%), Gaps = 30/431 (6%)
Query: 292 VIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVR 351
++DSGAS I S + + V A + + I IGD+ + T ++ V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI-KVL 90
Query: 352 HVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMI---- 407
H P I +L+S+ +L F + + G V+A K G Y V+++ ++
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 408 ---AVTEAINSSS-------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG-------V 450
+ S S H+ L H + + ++ ++ D+
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQ 209
Query: 451 CEHCILGKQRKVSFSKAGR---KSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTR 507
C C++GK K K R ++ E + +HTD++GP Y+++F D++T+
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 508 KVWVYFLKSKSD--VFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIR 565
WVY L + + + VF ++NQ + ++ D G EY ++ KF +NGI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 566 MIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLD 625
T +GVAER+NRTL + R SGLP W AI + ++ + P
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKS 388
Query: 626 YQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWD 685
+ + + +S L FG ++ D + K+ P+ I + + + YGY +
Sbjct: 389 KKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYL 447
Query: 686 EQNRKIIRSIN 696
+K + + N
Sbjct: 448 PSLKKTVDTTN 458
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 112 bits (280), Expect = 3e-24
Identities = 101/431 (23%), Positives = 180/431 (41%), Gaps = 30/431 (6%)
Query: 292 VIDSGASFHTIPSKELLSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVR 351
++DSGAS I S + + V A + + I IGD+ + T ++ V
Sbjct: 32 LLDSGASRTLIRSAHHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI-KVL 90
Query: 352 HVPGIKRNLISIGQLDDEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMI---- 407
H P I +L+S+ +L F + + G V+A K G Y V+++ ++
Sbjct: 91 HTPNIAYDLLSLNELAAVDITACFTKNVLERSDGT-VLAPIVKYGDFYWVSKKYLLPSNI 149
Query: 408 ---AVTEAINSSS-------IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG-------V 450
+ S S H+ L H + + ++ ++ D+
Sbjct: 150 SVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDRSSAIDYQ 209
Query: 451 CEHCILGKQRKVSFSKAGR---KSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTR 507
C C++GK K K R ++ E + +HTD++GP Y+++F D++T+
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 508 KVWVYFLKSKSD--VFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIR 565
WVY L + + + VF ++NQ + ++ D G EY ++ KF +NGI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 566 MIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLD 625
T +GVAER+NRTL + R SGLP W AI + ++ + P
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKS 388
Query: 626 YQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWD 685
+ + + +S L FG ++ D + K+ P+ I + + + YGY +
Sbjct: 389 KKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYL 447
Query: 686 EQNRKIIRSIN 696
+K + + N
Sbjct: 448 PSLKKTVDTTN 458
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 111 bits (277), Expect = 8e-24
Identities = 139/655 (21%), Positives = 249/655 (37%), Gaps = 54/655 (8%)
Query: 80 DLMKALSNMYEKPFA-ANKVHLIRRLFNLRMGEGNSVTEHINSFNTIISQLSSVKITFDN 138
D+MK LS EK + + + I L NL+ II +L++ I +N
Sbjct: 247 DIMKILSKSIEKMQSDTQEANDIVTLANLQYNGSTPADAFETKVTNIIDRLNNNGIHINN 306
Query: 139 ELMVLSLLQSLPDSWAATV--------TAVSNSARDNKLKFDDIRDLILSEDIRRKDSGE 190
++ +++ L + V+ D +++ + S+ R++ +
Sbjct: 307 KVACQLIMRGLSGEYKFLRYTRHRHLNMTVAELFLDIHAIYEEQQGSRNSKPNYRRNLSD 366
Query: 191 SSNTFGSALNTESR---GRGSQKSHNQSQGRGRSKSRGRSQTRVRNDITCWNCDRKGHFT 247
N S NT R QK++N R+ + S D +
Sbjct: 367 EKNDSRSYTNTTKPKVIARNPQKTNNSKSKTARAHNVSTSNNSPSTD--------NDSIS 418
Query: 248 NQCKAPRKKKNYQKR*DDDESANAATEEVADTLICSLDSPVDSWVIDSGASFHTIPSKEL 307
P + N D TE + S D ++DSGAS I S
Sbjct: 419 KSTTEPIQLNNKH----DLTLGQELTESTVNHTNHSDDELPGHLLLDSGASRTLIRSAHH 474
Query: 308 LSNYICGKFGKVYLADGKPLDIVGIGDIDIRSSNGTLWTLHNVRHVPGIKRNLISIGQLD 367
+ + V A + + I IGD+ + T ++ V H P I +L+S+ +L
Sbjct: 475 IHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSI-KVLHTPNIAYDLLSLNELA 533
Query: 368 DEGYHTTFGGGAWKVTKGNLVVARGKKRGSLYMVAEEDMI-------AVTEAINSSS--- 417
F + + G V+A + G Y V++ ++ + S S
Sbjct: 534 AVDITACFTKNVLERSDGT-VLAPIVQYGDFYWVSKRYLLPSNISVPTINNVHTSESTRK 592
Query: 418 ----IWHQRLGHMSEKGMKIMASKGKMSNLKHVDLG-------VCEHCILGKQRKVSFSK 466
H+ L H + + ++ ++ D+ C C++GK K K
Sbjct: 593 YPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIK 652
Query: 467 AGR---KSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKVWVYFLKSKSD--VF 521
R ++ E + +HTD++GP Y+++F D++T+ WVY L + + +
Sbjct: 653 GSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSIL 712
Query: 522 SVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAE 581
VF ++NQ + ++ D G EY ++ KF +NGI T +GVAE
Sbjct: 713 DVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAE 772
Query: 582 RMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLS 641
R+NRTL + R SGLP W AI + ++ + P + + + +S
Sbjct: 773 RLNRTLLDDCRTQLQCSGLPNHLWFSAIEFST-IVRNSLASPKSKKSARQHAGLAGLDIS 831
Query: 642 HLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSDMYGYRFWDEQNRKIIRSIN 696
L FG ++ D + K+ P+ I + + + YGY + +K + + N
Sbjct: 832 TLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTN 885
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 79.7 bits (195), Expect = 2e-14
Identities = 43/117 (36%), Positives = 66/117 (55%), Gaps = 7/117 (5%)
Query: 378 GAWKVTKGNLVVARGKKRGSLYMV-----AEEDMIAVTEAINSSSIWHQRLGHMSEKGMK 432
G KV KG + +G + SLY++ E +A T A + + +WH RL HMS++GM+
Sbjct: 27 GVLKVLKGCRTILKGNRHDSLYILQGSVETGESNLAET-AKDETRLWHSRLAHMSQRGME 85
Query: 433 IMASKGKMSNLKHVDLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHTDVWGPAPV 489
++ KG + + K L CE CI GK +V+FS G+ + L+ VH+D+WG V
Sbjct: 86 LLVKKGFLDSSKVSSLKFCEDCIYGKTHRVNFS-TGQHTTKNPLDYVHSDLWGAPSV 141
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 79.3 bits (194), Expect = 3e-14
Identities = 103/462 (22%), Positives = 183/462 (39%), Gaps = 39/462 (8%)
Query: 292 VIDSGASFHTIPSKELLSNYICGKFGKVYLADGK--PLDIVGIGDIDIRSS-NGTLWTLH 348
+ID+G+ + K LL NY + GK + + G G I I++ N T
Sbjct: 413 IIDTGSGVNITNDKTLLHNYEDSNRSTRFFGIGKNSSVSVKGYGYIKIKNGHNNTDNKCL 472
Query: 349 NVRHVPGIKRNLISIGQLDDEGY------HTTFGGGAWKV-TKGNLVVARGKKRGSLYMV 401
+VP + +IS L + +T G K+ TK V K +
Sbjct: 473 LTYYVPEEESTIISCYDLAKKTKMVLSRKYTRLGNKIIKIKTKIVNGVIHVKMNELIERP 532
Query: 402 AEEDMIAVTEA-------INSSSIW----HQRLGHMS----EKGMKIMASKGKMSNLKHV 446
+++ I + +N SI H+R+GH E +K + + +K
Sbjct: 533 SDDSKINAIKPTSSPGFKLNKRSITLEDAHKRMGHTGIQQIENSIKHNHYEESLDLIKEP 592
Query: 447 DLGVCEHCILGKQRKVSFSKAGRKSKSEKLELVHT---DVWGPAPVKSLGGSRYYVTFID 503
+ C+ C + K K + + S E + D++GP + RY + +D
Sbjct: 593 NEFWCQTCKISKATKRNHYTGSMNNHSTDHEPGSSWCMDIFGPVSSSNADTKRYMLIMVD 652
Query: 504 DSTRKVWV--YFLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSE 561
++TR +F K+ + + +K VE Q K++ + SD G E+ + + +++
Sbjct: 653 NNTRYCMTSTHFNKNAETILAQVRKNIQYVETQFDRKVREINSDRGTEFTNDQIEEYFIS 712
Query: 562 NGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAA----YLIN 617
GI I T NG AER RT+ A + QS L FW A+ +A YL +
Sbjct: 713 KGIHHILTSTQDHAANGRAERYIRTIITDATTLLRQSNLRVKFWEYAVTSATNIRNYLEH 772
Query: 618 RGPSVPLDYQLPEEVWYGKEVSLSHLKVFGCVSYVLIDSDKRDKLDPKAIKCFFIGYGSD 677
+ +LP + + V++ + +I + KL P + + +
Sbjct: 773 KSTG-----KLPLKAISRQPVTVRLMSFLPFGEKGIIWNHNHKKLKPSGLPSIILCKDPN 827
Query: 678 MYGYRFWDEQNRKIIRSINVTFNESVLYKDRSSAESMSSSKQ 719
YGY+F+ KI+ S N T + + ++++ S Q
Sbjct: 828 SYGYKFFIPSKNKIVTSDNYTIPNYTMDGRVRNTQNINKSHQ 869
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 78.6 bits (192), Expect = 5e-14
Identities = 38/86 (44%), Positives = 56/86 (64%), Gaps = 3/86 (3%)
Query: 583 MNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQLPEEVWYGKEVSLSH 642
MNRT+ E+ R M + GLPK F DA NTA ++IN+ PS +++ +P+EVW+ + S+
Sbjct: 1 MNRTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSY 60
Query: 643 LKVFGCVSYVLIDSDKRDKLDPKAIK 668
L+ FGCV+Y+ D KL P+A K
Sbjct: 61 LRRFGCVAYIHCD---EGKLKPRAKK 83
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 48.5 bits (114), Expect = 6e-05
Identities = 59/252 (23%), Positives = 103/252 (40%), Gaps = 13/252 (5%)
Query: 393 KKRGSLYMVAEEDMIAVTEAINSSSIWHQRLGHMSEKGMKIMASKGKMSNLKHVDLGVCE 452
+K G + + +E + + + + + +Q+L + EK ++ G + VC+
Sbjct: 682 EKEGKIVLPRKEALAMIQQMHAWTHLSNQKLKLLIEKTDFLIPKAGTLIEQVTSACKVCQ 741
Query: 453 HCILGKQRKVSFSKAGRKSKSEKLELVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKVWVY 512
G R + E+ T+V G +Y + F+D + V Y
Sbjct: 742 QVNAGATRVPEGKRTRGNRPGVYWEIDFTEV-----KPHYAGYKYLLVFVDTFSGWVEAY 796
Query: 513 FLKSKSDVFSVFKKWKTEVENQTGLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPG 572
+ ++ V KK E+ + GL K + SDNG + SQ + GI
Sbjct: 797 PTRQET-AHMVAKKILEEIFPRFGLP-KVIGSDNGPAFVSQVSQGLARTLGINWKLHCAY 854
Query: 573 TPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQL-PEE 631
P+ +G ERMNRT+ E + +++GL W ++ A + R + P + L P E
Sbjct: 855 RPQSSGQVERMNRTIKETLTKLTLETGLKD--WRRLLSLA---LLRARNTPNRFGLTPYE 909
Query: 632 VWYGKEVSLSHL 643
+ YG LS L
Sbjct: 910 ILYGGPPPLSTL 921
>POL_SMSAV (P03359) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 294
Score = 47.4 bits (111), Expect = 1e-04
Identities = 42/144 (29%), Positives = 65/144 (44%), Gaps = 14/144 (9%)
Query: 494 GSRYYVTFIDDSTRKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKI-KSLKSDNGGEYDS 552
G+RY + FID T WV +K++ K N T L+I K L SDNG + +
Sbjct: 26 GNRYLLVFID--TFSGWVEAFPTKTETALTVCK-----RNSTPLRIPKVLGSDNGPAFVA 78
Query: 553 QEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTA 612
Q + ++ GI P+ +G ERMNRT+ E + +++G D +
Sbjct: 79 QVSQGLATQLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLALETG-----GKDWVALL 133
Query: 613 AYLINRGPSVPLDYQL-PEEVWYG 635
+ R + P + L P E+ YG
Sbjct: 134 PLALLRAKNTPSRFGLTPYEILYG 157
>POL_MPMV (P07572) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 867
Score = 47.0 bits (110), Expect = 2e-04
Identities = 25/79 (31%), Positives = 43/79 (53%), Gaps = 1/79 (1%)
Query: 540 KSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTLNERARCMRIQSG 599
K +K+DNG Y S+ F++FCS I+ I IP P+ G+ ER + +L ++
Sbjct: 709 KQIKTDNGPGYTSKNFQEFCSTLQIKHITGIPYNPQGQGIVERAHLSLKTTIEKIKKGEW 768
Query: 600 LPKMFWP-DAINTAAYLIN 617
P+ P + +N A +++N
Sbjct: 769 YPRKGTPRNILNHALFILN 787
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 47.0 bits (110), Expect = 2e-04
Identities = 45/190 (23%), Positives = 75/190 (38%), Gaps = 13/190 (6%)
Query: 448 LGVCEHCILGKQRKVSFSKAGRKSKSEK-LELVHTDVWGPAPVKSLGGSRYYVTFIDDST 506
LG C+ C++ + R + +K + D GP P G Y + +D T
Sbjct: 647 LGRCQQCLITNASNKASGPILRPDRPQKPFDKFFIDYIGPLPPSQ--GYLYVLVVVDGMT 704
Query: 507 RKVWVYFLKSKSDVFSVFKKWKTEVENQTGLKI-KSLKSDNGGEYDSQEFKKFCSENGIR 565
W+Y K+ S +V + T + I K + SD G + S F ++ E GI
Sbjct: 705 GFTWLYPTKAPSTSATV-----KSLNVLTSIAIPKVIHSDQGAAFTSSTFAEWAKERGIH 759
Query: 566 MIKTIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLD 625
+ + P P+ ER N + + ++ G P W D + +N S L
Sbjct: 760 LEFSTPYHPQSGSKVERKNSDI--KRLLTKLLVGRPTK-WYDLLPVVQLALNNTYSPVLK 816
Query: 626 YQLPEEVWYG 635
Y P ++ +G
Sbjct: 817 Y-TPHQLLFG 825
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 46.6 bits (109), Expect = 2e-04
Identities = 45/187 (24%), Positives = 77/187 (41%), Gaps = 13/187 (6%)
Query: 451 CEHCILGKQRKVSFSKAGRKSKSEK-LELVHTDVWGPAPVKSLGGSRYYVTFIDDSTRKV 509
C+ C++ ++ R K K + + D GP P + G + + +D T V
Sbjct: 858 CKQCLVTNATNLTSPPILRPVKPLKPFDKFYIDYIGPLPPSN--GYLHVLVVVDSMTGFV 915
Query: 510 WVYFLKSKSDVFSVFKKWKTEVENQTGLKI-KSLKSDNGGEYDSQEFKKFCSENGIRMIK 568
W+Y K+ S +V + T + I K L SD G + S F + E GI++
Sbjct: 916 WLYPTKAPSTSATV-----KALNMLTSIAIPKVLHSDQGAAFTSSTFADWAKEKGIQLEF 970
Query: 569 TIPGTPEQNGVAERMNRTLNERARCMRIQSGLPKMFWPDAINTAAYLINRGPSVPLDYQL 628
+ P P+ +G ER N + + ++ G P W D + +N S P
Sbjct: 971 STPYHPQSSGKVERKNSDI--KRLLTKLLIGRPAK-WYDLLPVVQLALNNSYS-PSSKYT 1026
Query: 629 PEEVWYG 635
P ++ +G
Sbjct: 1027 PHQLLFG 1033
>POL_IPMAI (P12894) Probable Pol polyprotein [Contains: Integrase
(IN); Reverse transcriptase/ribonuclease H (EC 2.7.7.49)
(EC 3.1.26.4) (RT)]
Length = 814
Score = 46.6 bits (109), Expect = 2e-04
Identities = 20/50 (40%), Positives = 32/50 (64%)
Query: 538 KIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTL 587
K + LK+DNG Y SQ+F++FC + + + +P P+ G+ ER +RTL
Sbjct: 622 KPRLLKTDNGPAYTSQKFQQFCRQMDVTHLTGLPYNPQGQGIVERAHRTL 671
>POL_IPMA (P11368) Putative Pol polyprotein [Contains: Integrase
(IN); Reverse transcriptase/ribonuclease H (EC 2.7.7.49)
(EC 3.1.26.4) (RT)]
Length = 867
Score = 46.6 bits (109), Expect = 2e-04
Identities = 20/50 (40%), Positives = 32/50 (64%)
Query: 538 KIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTL 587
K + LK+DNG Y SQ+F++FC + + + +P P+ G+ ER +RTL
Sbjct: 704 KPRLLKTDNGPAYTSQKFQQFCRQMDVTHLTGLPYNPQGQGIVERAHRTL 753
>IGEB_MOUSE (P03975) IgE-binding protein
Length = 557
Score = 46.6 bits (109), Expect = 2e-04
Identities = 20/50 (40%), Positives = 32/50 (64%)
Query: 538 KIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTIPGTPEQNGVAERMNRTL 587
K + LK+DNG Y SQ+F++FC + + + +P P+ G+ ER +RTL
Sbjct: 434 KPRLLKTDNGPAYTSQKFQQFCRQMDVTHLTGLPYNPQGQGIVERAHRTL 483
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 83,812,933
Number of Sequences: 164201
Number of extensions: 3612710
Number of successful extensions: 10309
Number of sequences better than 10.0: 211
Number of HSP's better than 10.0 without gapping: 161
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 9904
Number of HSP's gapped (non-prelim): 376
length of query: 720
length of database: 59,974,054
effective HSP length: 118
effective length of query: 602
effective length of database: 40,598,336
effective search space: 24440198272
effective search space used: 24440198272
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0220.7


