

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
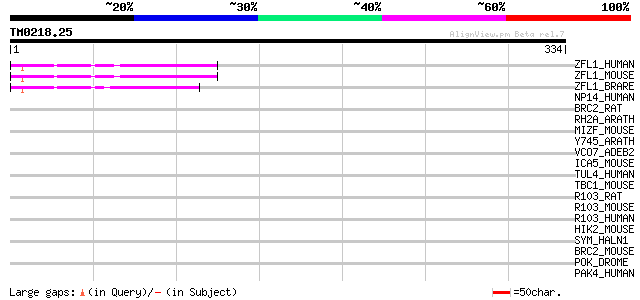
Query= TM0218.25
(334 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ZFL1_HUMAN (O95159) Zinc finger protein-like 1 (Zinc-finger prot... 95 3e-19
ZFL1_MOUSE (Q9DB43) Zinc finger protein-like 1 94 4e-19
ZFL1_BRARE (P62447) Zinc finger protein-like 1 92 2e-18
NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130... 36 0.15
BRC2_RAT (O35923) Breast cancer type 2 susceptibility protein ho... 35 0.20
RH2A_ARATH (Q9ZT50) RING-H2 zinc finger protein RHA2a 34 0.59
MIZF_MOUSE (Q8K1K9) MBD2-interacting zinc finger protein (Methyl... 34 0.59
Y745_ARATH (P59278) Hypothetical protein At1g51745 33 0.77
VCO7_ADEB2 (Q96624) Major core protein precursor (Protein VII) (... 33 1.3
ICA5_MOUSE (Q60625) Intercellular adhesion molecule-5 precursor ... 33 1.3
TUL4_HUMAN (Q9NRJ4) Tubby-like protein 4 (Tubby superfamily prot... 32 1.7
TBC1_MOUSE (Q60949) TBC1 domain family member 1 32 2.2
R103_RAT (Q9EPZ8) RING finger protein 103 (Zinc finger protein 1... 32 2.2
R103_MOUSE (Q9R1W3) RING finger protein 103 (Zinc finger protein... 32 2.2
R103_HUMAN (O00237) RING finger protein 103 (Zinc finger protein... 32 2.2
HIK2_MOUSE (Q9QZR5) Homeodomain-interacting protein kinase 2 (EC... 32 2.2
SYM_HALN1 (Q9HSA4) Methionyl-tRNA synthetase (EC 6.1.1.10) (Meth... 32 2.9
BRC2_MOUSE (P97929) Breast cancer type 2 susceptibility protein ... 32 2.9
POK_DROME (Q01842) Ets DNA-binding protein pokkuri (Protein yan)... 31 5.0
PAK4_HUMAN (O96013) Serine/threonine-protein kinase PAK 4 (EC 2.... 31 5.0
>ZFL1_HUMAN (O95159) Zinc finger protein-like 1 (Zinc-finger protein
MCG4)
Length = 310
Score = 94.7 bits (234), Expect = 3e-19
Identities = 50/128 (39%), Positives = 70/128 (54%), Gaps = 9/128 (7%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKRKVTNLFCFEHRVNVCEHCLV-ANHAKCIVQSYLQWLQDSDYN--PNCRLCN 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDS- 117
L +TTRL C + H CL P +TAPAGY CP+C+ I+PP ++
Sbjct: 58 IPLAS---RETTRLVCYDLFHWACLNERAAQLPRNTAPAGYQCPSCNGPIFPPTNLAGPV 114
Query: 118 GSRLHSKL 125
S L KL
Sbjct: 115 ASALREKL 122
>ZFL1_MOUSE (Q9DB43) Zinc finger protein-like 1
Length = 310
Score = 94.4 bits (233), Expect = 4e-19
Identities = 49/128 (38%), Positives = 70/128 (54%), Gaps = 9/128 (7%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKRKVTNLFCFEHRVNVCEHCLV-ANHAKCIVQSYLQWLQDSDYN--PNCRLCN 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDS- 117
L +TTRL C + H C+ P +TAPAGY CP+C+ I+PP ++
Sbjct: 58 TPLAS---RETTRLVCYDLFHWACINERAAQLPRNTAPAGYQCPSCNGPIFPPANLAGPV 114
Query: 118 GSRLHSKL 125
S L KL
Sbjct: 115 ASALREKL 122
>ZFL1_BRARE (P62447) Zinc finger protein-like 1
Length = 317
Score = 92.0 bits (227), Expect = 2e-18
Identities = 43/116 (37%), Positives = 66/116 (56%), Gaps = 8/116 (6%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC +K T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKKKVTNLFCFKHRVNVCEHCLV-SNHNKCIVQSYLQWLQDSDYN--PNCSLC- 56
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSV 114
++ D T RL C + H +CL P +TAP GY CPTC ++PP+++
Sbjct: 57 --IQPLDSQDTVRLVCYDLFHWSCLNELASHQPLNTAPDGYQCPTCQGPVFPPRNL 110
>NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130
kDa protein) (140 kDa nucleolar phosphoprotein)
(Nopp140) (Nucleolar and coiled-body phosphoprotein 1)
Length = 699
Score = 35.8 bits (81), Expect = 0.15
Identities = 48/200 (24%), Positives = 76/200 (38%), Gaps = 16/200 (8%)
Query: 102 PTCSTSIWPPKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASE 161
P + PPK K S S S ++ + K PV++ P PA A+
Sbjct: 152 PQAKAAKAPPKKAKSSDSDSDSSSEDEPPKNQKPKIT----PVTVKAQTKAPPKPARAAP 207
Query: 162 PLIGRENHGNSDSPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSPVGPATRKGAFNVER 221
+ + +S S ++ S S D E + A + V V A K A R
Sbjct: 208 KIANGKAASSSSSSSSSS-----SSDDSEEEKAAATPKKTVPKKQVVAKAPVKAATTPTR 262
Query: 222 QNSEISYYADDEDGNRKKYTKRGPFYHKFLRALLPFWSSALPTLPV-TAPPRK--DASNA 278
++S + DE+ +KK K P + + P S+ P + T PP+K +
Sbjct: 263 KSSSSEDSSSDEEEEQKKPMKNKPGPYSY----APPPSAPPPKKSLGTQPPKKAVEKQQP 318
Query: 279 TEGSEGRTRHQRSSRMDPRK 298
E SE + SS + +K
Sbjct: 319 VESSEDSSDESDSSSEEEKK 338
>BRC2_RAT (O35923) Breast cancer type 2 susceptibility protein
homolog
Length = 3343
Score = 35.4 bits (80), Expect = 0.20
Identities = 23/92 (25%), Positives = 39/92 (42%), Gaps = 2/92 (2%)
Query: 217 FNVERQNSEISYYADDEDGNRKKYTKRGPFYHKFLRALLPFWSSALPTLPVTAPPRKDAS 276
+N E E Y + K +R P YH+F + F + TLP+ P K+
Sbjct: 42 YNTEHPE-ESEYKPQGHEPQLFKTPQRNPSYHQFASTPIMFKEQS-QTLPLDQSPFKELG 99
Query: 277 NATEGSEGRTRHQRSSRMDPRKILLLIAIMAC 308
N S+ + ++ +R DP + + + AC
Sbjct: 100 NVVANSKRKHHSKKKARKDPVVDVASLPLKAC 131
>RH2A_ARATH (Q9ZT50) RING-H2 zinc finger protein RHA2a
Length = 155
Score = 33.9 bits (76), Expect = 0.59
Identities = 16/55 (29%), Positives = 26/55 (47%), Gaps = 9/55 (16%)
Query: 54 CCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSI 108
C C + L+EG+ + +L C HV H CL + F + CP C +++
Sbjct: 86 CVVCLSKLKEGE--EVRKLECRHVFHKKCLEGWLHQF-------NFTCPLCRSAL 131
>MIZF_MOUSE (Q8K1K9) MBD2-interacting zinc finger protein
(Methyl-CpG-binding protein 2-interacting zinc finger
protein)
Length = 503
Score = 33.9 bits (76), Expect = 0.59
Identities = 26/83 (31%), Positives = 35/83 (41%), Gaps = 11/83 (13%)
Query: 29 PEHQICVIRTYSEWVIDG------EYDWPPKCCQCQAVLEEGDGSQTTRLGCLHVIHTNC 82
P+H +C+ + E V D D CC+ QAV ++ Q GC C
Sbjct: 122 PDHFLCLWE-HCESVFDNPEWFYRHVDAHSLCCEYQAVSKDNHVVQCGWKGCTCTFKDRC 180
Query: 83 -LVSHIKSFPPHTAPAGYACPTC 104
L H++S HT ACPTC
Sbjct: 181 KLREHLRS---HTQEKVVACPTC 200
>Y745_ARATH (P59278) Hypothetical protein At1g51745
Length = 597
Score = 33.5 bits (75), Expect = 0.77
Identities = 24/68 (35%), Positives = 28/68 (40%), Gaps = 1/68 (1%)
Query: 145 SLSVTESRSPPPAFASEPLIGRENHGNSDSPATG-SEPPKLSVTDIVEIEGANSAGNFVK 203
S+SV+ PL G ENH A S P K VTD+ G NS FVK
Sbjct: 357 SISVSAEDDSSDRLFDVPLTGEENHSEGFPAACRISSPRKALVTDLTRRCGRNSHNVFVK 416
Query: 204 GSSPVGPA 211
+ G A
Sbjct: 417 NEASNGSA 424
>VCO7_ADEB2 (Q96624) Major core protein precursor (Protein VII)
(pVII)
Length = 183
Score = 32.7 bits (73), Expect = 1.3
Identities = 21/64 (32%), Positives = 32/64 (49%), Gaps = 2/64 (3%)
Query: 260 SALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIMACMATMGILYYRL 319
+ +PT+P+T P D NA +EG TR +R + R+ A+ A A + RL
Sbjct: 50 TTVPTVPITDDPVADVVNAI--AEGTTRRRRRAERRRRRRQATSAMRAARALVRSARRRL 107
Query: 320 VQRG 323
+RG
Sbjct: 108 ARRG 111
>ICA5_MOUSE (Q60625) Intercellular adhesion molecule-5 precursor
(ICAM-5) (Telencephalin)
Length = 917
Score = 32.7 bits (73), Expect = 1.3
Identities = 16/59 (27%), Positives = 25/59 (42%), Gaps = 4/59 (6%)
Query: 56 QCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSV 114
+C A G S+T +G + +V+ + + PP P G TC WPP +
Sbjct: 723 RCVATNAHGTDSRTVTVG----VEYRPVVAELAASPPSVRPGGNFTLTCRAEAWPPAQI 777
>TUL4_HUMAN (Q9NRJ4) Tubby-like protein 4 (Tubby superfamily protein)
Length = 1544
Score = 32.3 bits (72), Expect = 1.7
Identities = 32/129 (24%), Positives = 55/129 (41%), Gaps = 6/129 (4%)
Query: 93 HTAPAGYACPTCSTSIW-PPKSVKDSGSRLHSKLKE--AIMQ-TGMEKNIFGNHPVSLSV 148
HTA A S S+ PP S +D ++S E A+ Q +EK + HP
Sbjct: 1056 HTASASPLASQSSYSLLSPPDSARDRTDYVNSAFTEDEALSQHCQLEKPL--RHPPLPEA 1113
Query: 149 TESRSPPPAFASEPLIGRENHGNSDSPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSPV 208
+ PP + +P++G + + A S P L ++ ++ +G + + + SP
Sbjct: 1114 AVTLKRPPPYQWDPMLGEDVWVPQERTAQTSGPNPLKLSSLMLSQGQHLDVSRLPFISPK 1173
Query: 209 GPATRKGAF 217
PA+ F
Sbjct: 1174 SPASPTATF 1182
>TBC1_MOUSE (Q60949) TBC1 domain family member 1
Length = 1255
Score = 32.0 bits (71), Expect = 2.2
Identities = 41/190 (21%), Positives = 67/190 (34%), Gaps = 9/190 (4%)
Query: 76 HVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSRLHSKLKEAIMQTGME 135
H+ + L+S ++F + C P +S R + +T E
Sbjct: 570 HIAEESALLSPQQAFRRRANTLSHFPVECPAPPEPAQSSPGVSQRKLMRYHSVSTETPHE 629
Query: 136 KNIF---GNHPVSLSVTESRSPPPAF---ASEPLIGRENHGNSDSPATGSEPPK-LSVTD 188
+N+ G S PPP AS P + NS +G+ PK +S +
Sbjct: 630 RNVDHLPGGESQGCPGQPSAPPPPRLNPSASSPNFFKYLKHNSSGEQSGNAVPKSVSYRN 689
Query: 189 IV--EIEGANSAGNFVKGSSPVGPATRKGAFNVERQNSEISYYADDEDGNRKKYTKRGPF 246
+ ++ ++S NF+K +PV N R + + D DG K +
Sbjct: 690 ALRKKLHSSSSVPNFLKFLAPVDENNTCDFKNTNRDFESKANHLGDTDGTPVKTRRHSWR 749
Query: 247 YHKFLRALLP 256
FLR P
Sbjct: 750 QQIFLRVATP 759
>R103_RAT (Q9EPZ8) RING finger protein 103 (Zinc finger protein 103)
(Zfp-103) (ADRG34 protein)
Length = 682
Score = 32.0 bits (71), Expect = 2.2
Identities = 21/65 (32%), Positives = 26/65 (39%), Gaps = 14/65 (21%)
Query: 50 WPP---KCCQCQAVLEEGD-GSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCS 105
WP C +C LE + G L C HV H NC+V + A + CP C
Sbjct: 608 WPAGTLHCTECVVCLENFENGCLLMGLPCGHVFHQNCIVMWL-------AGGRHCCPVCR 660
Query: 106 TSIWP 110
WP
Sbjct: 661 ---WP 662
>R103_MOUSE (Q9R1W3) RING finger protein 103 (Zinc finger protein
103) (Zfp-103) (KF-1) (mKF-1)
Length = 683
Score = 32.0 bits (71), Expect = 2.2
Identities = 21/65 (32%), Positives = 26/65 (39%), Gaps = 14/65 (21%)
Query: 50 WPP---KCCQCQAVLEEGD-GSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCS 105
WP C +C LE + G L C HV H NC+V + A + CP C
Sbjct: 609 WPAGTLHCTECVVCLENFENGCLLMGLPCGHVFHQNCIVMWL-------AGGRHCCPVCR 661
Query: 106 TSIWP 110
WP
Sbjct: 662 ---WP 663
>R103_HUMAN (O00237) RING finger protein 103 (Zinc finger protein
103 homolog) (Zfp-103) (KF-1) (hKF-1)
Length = 685
Score = 32.0 bits (71), Expect = 2.2
Identities = 21/65 (32%), Positives = 26/65 (39%), Gaps = 14/65 (21%)
Query: 50 WPPK---CCQCQAVLEEGD-GSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCS 105
WP C +C LE + G L C HV H NC+V + A + CP C
Sbjct: 611 WPADMLHCTECVVCLENFENGCLLMGLPCGHVFHQNCIVMWL-------AGGRHCCPVCR 663
Query: 106 TSIWP 110
WP
Sbjct: 664 ---WP 665
>HIK2_MOUSE (Q9QZR5) Homeodomain-interacting protein kinase 2 (EC
2.7.1.-) (Nuclear body associated kinase 1) (Sialophorin
tail associated nuclear serine/threonine kinase)
Length = 1196
Score = 32.0 bits (71), Expect = 2.2
Identities = 23/83 (27%), Positives = 35/83 (41%), Gaps = 7/83 (8%)
Query: 83 LVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNH 142
+ SH++ F PHT + C + P + +G HSK+ KNI +
Sbjct: 8 MASHVQVFSPHTLQSSAFCSVKKLKVEPSSNWDMTGYGSHSKV------YSQSKNIPPSQ 61
Query: 143 PVSLSVTESRS-PPPAFASEPLI 164
P S +V+ S P P+ E I
Sbjct: 62 PASTTVSTSLPIPNPSLPYEQTI 84
>SYM_HALN1 (Q9HSA4) Methionyl-tRNA synthetase (EC 6.1.1.10)
(Methionine--tRNA ligase) (MetRS)
Length = 689
Score = 31.6 bits (70), Expect = 2.9
Identities = 24/80 (30%), Positives = 35/80 (43%), Gaps = 15/80 (18%)
Query: 29 PEHQICVIRTYSEWVIDGEYDWPPKCCQCQAVLEEGDGSQTTRLGCLHVIHTNCLVSHIK 88
P + + YSE V E+DW +V ++GDG +HVI + + H
Sbjct: 263 PIEYVAATKQYSERVGTEEFDW-------ASVWKDGDGE------IIHVIGRDIIQHHTV 309
Query: 89 SFPPHTAPAGYACP--TCST 106
+P A A YA P C+T
Sbjct: 310 FWPSMLAGADYAEPRAVCAT 329
>BRC2_MOUSE (P97929) Breast cancer type 2 susceptibility protein
homolog
Length = 3329
Score = 31.6 bits (70), Expect = 2.9
Identities = 31/117 (26%), Positives = 51/117 (43%), Gaps = 16/117 (13%)
Query: 104 CSTSIWPPKSVKDSGSRLHS-KLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEP 162
C+ ++ + SGS ++ KLKE + +++ +S T S + PP +S
Sbjct: 141 CTQAVLQREKPVVSGSLFYTPKLKEGQTPKPISESLGVEVDPDMSWTSSLATPPTLSSTV 200
Query: 163 LIGRENHGNS-----DSPAT-------GSEPPK---LSVTDIVEIEGANSAGNFVKG 204
LI R+ S DSPAT +E P+ SV +++ E N F +G
Sbjct: 201 LIARDEEARSSVTPADSPATLKSCFSNHNESPQKNDRSVPSVIDSENKNQQEAFSQG 257
Score = 30.4 bits (67), Expect = 6.5
Identities = 15/58 (25%), Positives = 29/58 (49%), Gaps = 1/58 (1%)
Query: 239 KYTKRGPFYHKFLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDP 296
K +R P YH+F + F + TLP+ P ++ S+ +T ++ +++DP
Sbjct: 63 KTPQRNPPYHQFASTPIMFKERS-QTLPLDQSPFRELGKVVASSKHKTHSKKKTKVDP 119
>POK_DROME (Q01842) Ets DNA-binding protein pokkuri (Protein yan)
(Protein anterior open)
Length = 732
Score = 30.8 bits (68), Expect = 5.0
Identities = 46/228 (20%), Positives = 79/228 (34%), Gaps = 31/228 (13%)
Query: 92 PHTAPAGYACPTCSTSIWPPKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTES 151
PH+ P T WPP + S HS M N P +++
Sbjct: 139 PHSHPP--------TPTWPPLNAPPENSPFHSSAHSLAGHHFMAPNSVTLSPPPSVDSQA 190
Query: 152 RSPPPAFASEPLIGRENHGNSDSPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSP-VGP 210
SPP A P G + A GS P T+ ++++ GS P + P
Sbjct: 191 SSPPQA----PYQNGGATGAAPGSAGGSAPAAGGATNTSNPTSSSASSTGSNGSQPNIMP 246
Query: 211 ATRKGAFNVERQNSEISYYADDEDGNRK------KYTKRGPFYHKFLRALLPFWSSALPT 264
+ + +SE Y++ G K Y+ P L+ + P W+ L
Sbjct: 247 MKGISSASSNHSDSE-EEYSETSGGVSKMPPAPLSYSTASPPGTPILKDIKPNWTQQLTN 305
Query: 265 LPVTAPPRKD-----------ASNATEGSEGRTRHQRSSRMDPRKILL 301
V + ++ A+ A + + + + Q+ + P+K+ L
Sbjct: 306 SFVNSWSQQQQQQQQQQAAAVAAVAAQAQQHQLQQQQQQQQLPQKLTL 353
>PAK4_HUMAN (O96013) Serine/threonine-protein kinase PAK 4 (EC
2.7.1.37) (p21-activated kinase 4) (PAK-4)
Length = 591
Score = 30.8 bits (68), Expect = 5.0
Identities = 47/188 (25%), Positives = 71/188 (37%), Gaps = 26/188 (13%)
Query: 102 PTCSTSIWP--PKSVKDSGSRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFA 159
P C TSI P PK++ R K+ + +++ F N V+ S + R PP
Sbjct: 56 PACITSIQPGAPKTIV----RGSKGAKDGALTLLLDE--FENMSVTRSNSLRRDSPPP-- 107
Query: 160 SEPLIGRENHGNSDSPATGSE--PPKLSVTDIVEIEGANSAGNFVKGS-SPVGPATRKGA 216
P R+ +G + PAT + P K A S G F S + G R+ A
Sbjct: 108 --PARARQENGMPEEPATTARGGPGK-----------AGSRGRFAGHSEAGGGSGDRRRA 154
Query: 217 FNVERQNSEISYYADDEDGNRKKYTKRGPFYHKFLRALLPFWSSALPTLPVTAPPRKDAS 276
+R S ++ +R K GP A L + P PR D
Sbjct: 155 GPEKRPKSSREGSGGPQESSRDKRPLSGPDVGTPQPAGLASGAKLAAGRPFNTYPRADTD 214
Query: 277 NATEGSEG 284
+ + G++G
Sbjct: 215 HPSRGAQG 222
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,561,416
Number of Sequences: 164201
Number of extensions: 1916536
Number of successful extensions: 4703
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 4684
Number of HSP's gapped (non-prelim): 39
length of query: 334
length of database: 59,974,054
effective HSP length: 111
effective length of query: 223
effective length of database: 41,747,743
effective search space: 9309746689
effective search space used: 9309746689
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0218.25


