

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
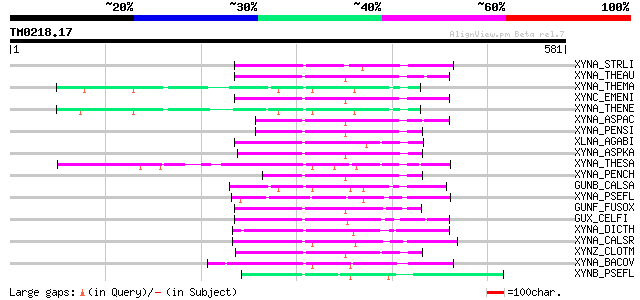
Query= TM0218.17
(581 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
XYNA_STRLI (P26514) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 114 7e-25
XYNA_THEAU (P23360) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8... 104 7e-22
XYNA_THEMA (Q60037) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 99 4e-20
XYNC_EMENI (Q00177) Endo-1,4-beta-xylanase C precursor (EC 3.2.1... 97 9e-20
XYNA_THENE (Q60042) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 97 1e-19
XYNA_ASPAC (O59859) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8... 97 1e-19
XYNA_PENSI (P56588) Endo-1,4-beta-xylanase (EC 3.2.1.8) (Xylanas... 94 1e-18
XLNA_AGABI (O60206) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8... 93 2e-18
XYNA_ASPKA (P33559) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 92 3e-18
XYNA_THESA (P36917) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 90 1e-17
XYNA_PENCH (P29417) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8... 90 1e-17
GUNB_CALSA (P10474) Endoglucanase/exoglucanase B precursor [Incl... 90 1e-17
XYNA_PSEFL (P14768) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 87 2e-16
GUNF_FUSOX (P46239) Putative endoglucanase type F precursor (EC ... 86 2e-16
GUX_CELFI (P07986) Exoglucanase/xylanase precursor [Includes: Ex... 84 1e-15
XYNA_DICTH (Q12603) Beta-1,4-xylanase (EC 3.2.1.8) (Endo-1,4-bet... 82 3e-15
XYNA_CALSR (P40944) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 80 1e-14
XYNZ_CLOTM (P10478) Endo-1,4-beta-xylanase Z precursor (EC 3.2.1... 80 1e-14
XYNA_BACOV (P49942) Endo-1,4-beta-xylanase A precursor (EC 3.2.1... 73 2e-12
XYNB_PSEFL (P23030) Endo-1,4-beta-xylanase B precursor (EC 3.2.1... 73 2e-12
>XYNA_STRLI (P26514) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 477
Score = 114 bits (285), Expect = 7e-25
Identities = 74/241 (30%), Positives = 114/241 (46%), Gaps = 25/241 (10%)
Query: 236 FGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNG 295
FG I + L++S Y F T N+MK +TE +GQ N+++ D + + A NG
Sbjct: 57 FGTAIASGRLSDSTYTSIAGREFNMVTAENEMKIDATEPQRGQFNFSSADRVYNWAVQNG 116
Query: 296 ISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRINSVASRYSGELIAWDVVNENLHFN 355
VRGH + W QQP W++SLS LR+A IN V + Y G+++ WDVVNE
Sbjct: 117 KQVRGHTLAWHS--QQPGWMQSLSGSALRQAMIDHINGVMAHYKGKIVQWDVVNE----A 170
Query: 356 FFEGNLGENASA-----------AYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYI 404
F +G+ G + + TA DP+ ++ N++N ++ + N +
Sbjct: 171 FADGSSGARRDSNLQRSGNDWIEVAFRTARAADPSAKLCYNDYNVENWTWAKTQAMYNMV 230
Query: 405 KKIQQILQFPGSAGIPL-AIGLQCHFASGQPNLAYMRAGLDLLGATGFPIWLTETSIDPQ 463
+ +Q G+P+ +G Q HF SG P + R L A G + +TE I
Sbjct: 231 RDFKQ-------RGVPIDCVGFQSHFNSGSPYNSNFRTTLQNFAALGVDVAITELDIQGA 283
Query: 464 P 464
P
Sbjct: 284 P 284
>XYNA_THEAU (P23360) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8)
(Xylanase) (1,4-beta-D-xylan xylanohydrolase) (TAXI)
Length = 329
Score = 104 bits (259), Expect = 7e-22
Identities = 71/234 (30%), Positives = 115/234 (48%), Gaps = 20/234 (8%)
Query: 236 FGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNG 295
FG + N LT + + F T N MKW +TE +G N+ D +++ A+ NG
Sbjct: 44 FGVATDQNRLTTGKNAAIIQADFGQVTPENSMKWDATEPSQGNFNFAGADYLVNWAQQNG 103
Query: 296 ISVRGHNIFWDDPKQQPQWVKSLSPEE-LRKAAAKRINSVASRYSGELIAWDVVNE---- 350
+RGH + W Q P WV S++ + L I ++ +RY G++ AWDVVNE
Sbjct: 104 KLIRGHTLVWH--SQLPSWVSSITDKNTLTNVMKNHITTLMTRYKGKIRAWDVVNEAFNE 161
Query: 351 --NLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQ 408
+L F +GE+ + TA DPN ++++N++N S + N +K+ +
Sbjct: 162 DGSLRQTVFLNVIGEDYIPIAFQTARAADPNAKLYINDYNLDSASYPKTQAIVNRVKQWR 221
Query: 409 QILQFPGSAGIPL-AIGLQCHFASGQPNLAYMRAGLDLLGATGFP-IWLTETSI 460
+AG+P+ IG Q H ++GQ A + L LL + G P + +TE +
Sbjct: 222 -------AAGVPIDGIGSQTHLSAGQG--AGVLQALPLLASAGTPEVAITELDV 266
>XYNA_THEMA (Q60037) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 1059
Score = 98.6 bits (244), Expect = 4e-20
Identities = 97/419 (23%), Positives = 166/419 (39%), Gaps = 77/419 (18%)
Query: 50 GGGIIVNPGFDHNLEGWTVFGNGAIEER----RSNEGNAFIVARNRTQPLDSFSQKIQLK 105
G +I F++ + W G+ IE S + + FI R + + K LK
Sbjct: 202 GPKVIYETSFENGVGDWQPRGDVNIEASSEVAHSGKSSLFISNRQKGWQGAQINLKGILK 261
Query: 106 KGMLYTFSAWLQLSEGSDTVSVV-------------FKINGRELIRGGHVIAKHGCWTLM 152
G Y F AW+ + G D ++ ++ + G + G +T+
Sbjct: 262 TGKTYAFEAWVYQNSGQDQTIIMTMQRKYSSDASTQYEWIKSATVPSGQWVQLSGTYTIP 321
Query: 153 KGGLVANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRSHQEESIERVRKSKVRFQV 212
G V + T + FES+NPT+E + D V + T + + E E +V
Sbjct: 322 AGVTVEDLT----LYFESQNPTLEFYVDDVKIVDTTSAEIKIEMEPEKEIPALKEV---- 373
Query: 213 THINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYST 272
K F G + + + N + + F T N+MK S
Sbjct: 374 -----------------LKDYFKVGVALPSKVFLNPKDIELITKHFNSITAENEMKPES- 415
Query: 273 EIVKGQEN------YTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWV------KSLSP 320
++ G EN + T D + + NG+ +RGH + W + Q P W LS
Sbjct: 416 -LLAGIENGKLKFRFETADKYIQFVEENGMVIRGHTLVWHN--QTPDWFFKDENGNLLSK 472
Query: 321 EELRKAAAKRINSVASRYSGELIAWDVVNENLHFNFFEG--------NLGENASAAYYST 372
E + + + I++V + G++ AWDVVNE + N +G +G + +
Sbjct: 473 EAMTERLKEYIHTVVGHFKGKVYAWDVVNEAVDPNQPDGLRRSTWYQIMGPDYIELAFKF 532
Query: 373 AHHLDPNTRMFLNEFNTIE-YSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCHFA 430
A DP+ ++F N++NT E D+ N +K +++ G+ IG+QCH +
Sbjct: 533 AREADPDAKLFYNDYNTFEPRKRDII---YNLVKDLKE-------KGLIDGIGMQCHIS 581
>XYNC_EMENI (Q00177) Endo-1,4-beta-xylanase C precursor (EC 3.2.1.8)
(Xylanase C) (1,4-beta-D-xylan xylanohydrolase C) (34
kDa xylanase) (Xylanase X34)
Length = 327
Score = 97.4 bits (241), Expect = 9e-20
Identities = 68/234 (29%), Positives = 106/234 (45%), Gaps = 18/234 (7%)
Query: 236 FGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNG 295
FG + +L NS+ + S+F T N MKW + E +G ++ D ++ A +
Sbjct: 41 FGTCSDQALLQNSQNEAIVASQFGVITPENSMKWDALEPSQGNFGWSGADYLVDYATQHN 100
Query: 296 ISVRGHNIFWDDPKQQPQWVKSL-SPEELRKAAAKRINSVASRYSGELIAWDVVNE---- 350
VRGH + W Q P WV S+ LR IN V RY G+++ WDVVNE
Sbjct: 101 KKVRGHTLVWH--SQLPSWVSSIGDANTLRSVMTNHINEVVGRYKGKIMHWDVVNEIFNE 158
Query: 351 --NLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQ 408
+ F LGE+ + TA DP+ ++++N++N S + +Y+KK
Sbjct: 159 DGTFRNSVFYNLLGEDFVRIAFETARAADPDAKLYINDYNLDSASYAKTQAMASYVKKWL 218
Query: 409 QILQFPGSAGIPL-AIGLQCHFASGQPNLAYMRAGLDLLGATGF-PIWLTETSI 460
+ G+P+ IG Q H++S + L L TG + +TE I
Sbjct: 219 -------AEGVPIDGIGSQAHYSSSHWSSTEAAGALSSLANTGVSEVAITELDI 265
>XYNA_THENE (Q60042) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
(Endoxylanase)
Length = 1055
Score = 97.1 bits (240), Expect = 1e-19
Identities = 96/422 (22%), Positives = 168/422 (39%), Gaps = 82/422 (19%)
Query: 50 GGGIIVNPGFDHNLEGWTVFGNG---AIEERRSNEGNAFIVARNRTQPLDS--FSQKIQL 104
G ++ F+ + W G+ +I + ++ G + NR + S K L
Sbjct: 197 GPKVVYETSFEKGIGDWQPRGSDVKISISPKVAHSGKKSLFVSNRQKGWHGAQISLKGIL 256
Query: 105 KKGMLYTFSAWLQLSEGSDTVSVV-------------FKINGRELIRGGHVIAKHGCWTL 151
K G Y F AW+ G D ++ ++ + G + G +T+
Sbjct: 257 KTGKTYAFEAWVYQESGQDQTIIMTMQRKYSSDSSTKYEWIKAATVPSGQWVQLSGTYTI 316
Query: 152 MKGGLVANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWR--SHQEESIERVRKSKVR 209
G V + T + FES+NPT+E + D V + T + + + EE I ++
Sbjct: 317 PAGVTVEDLT----LYFESQNPTLEFYVDDVKVVDTTSAEIKLEMNPEEEIPALKDVLKD 372
Query: 210 FQVTHINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKW 269
+ F G + + + N + +T N+MK
Sbjct: 373 Y-----------------------FRVGVALPSKVFINQKDIALISKHSNSSTAENEMK- 408
Query: 270 YSTEIVKGQEN------YTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWV------KS 317
++ G EN + T D + A+ NG+ VRGH + W + Q P+W
Sbjct: 409 -PDSLLAGIENGKLKFRFETADKYIEFAQQNGMVVRGHTLVWHN--QTPEWFFKDENGNL 465
Query: 318 LSPEELRKAAAKRINSVASRYSGELIAWDVVNENLHFNFFEG--------NLGENASAAY 369
LS EE+ + + I++V + G++ AWDVVNE + N +G +G +
Sbjct: 466 LSKEEMTERLREYIHTVVGHFKGKVYAWDVVNEAVDPNQPDGLRRSTWYQIMGPDYIELA 525
Query: 370 YSTAHHLDPNTRMFLNEFNTIE-YSGDVAASPTNYIKKIQQILQFPGSAGIPLAIGLQCH 428
+ A DPN ++F N++NT E D+ N +K +++ G+ IG+QCH
Sbjct: 526 FKFAREADPNAKLFYNDYNTFEPKKRDII---YNLVKSLKE-------KGLIDGIGMQCH 575
Query: 429 FA 430
+
Sbjct: 576 IS 577
>XYNA_ASPAC (O59859) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8)
(Xylanase) (1,4-beta-D-xylan xylanohydrolase)
(FIA-xylanase)
Length = 327
Score = 97.1 bits (240), Expect = 1e-19
Identities = 67/211 (31%), Positives = 107/211 (49%), Gaps = 18/211 (8%)
Query: 258 FKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKS 317
F T N MKW +TE +GQ +++ D +++ A++NG +RGH + W Q P WV+S
Sbjct: 66 FGQLTPENSMKWDATEPNRGQFSFSGSDYLVNFAQSNGKLIRGHTLVWH--SQLPSWVQS 123
Query: 318 LSPE-ELRKAAAKRINSVASRYSGELIAWDVVNE------NLHFNFFEGNLGENASAAYY 370
+ + L + I +V RY G++ AWDVVNE +L + F +GE+ +
Sbjct: 124 IYDKGTLIQVMQNHIATVMQRYKGKVYAWDVVNEIFNEDGSLRQSHFYNVIGEDYVRIAF 183
Query: 371 STAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPL-AIGLQCHF 429
TA +DPN ++++N++N S N++KK +AG+P+ IG Q H
Sbjct: 184 ETARAVDPNAKLYINDYNLDSASYPKLTGLVNHVKKWV-------AAGVPIDGIGSQTHL 236
Query: 430 ASGQPNLAYMRAGLDLLGATGFPIWLTETSI 460
++G A A L GA + +TE I
Sbjct: 237 SAG-AGAAVSGALNALAGAGTKEVAITELDI 266
>XYNA_PENSI (P56588) Endo-1,4-beta-xylanase (EC 3.2.1.8) (Xylanase)
(1,4-beta-D-xylan xylanohydrolase)
Length = 302
Score = 94.0 bits (232), Expect = 1e-18
Identities = 57/183 (31%), Positives = 94/183 (51%), Gaps = 17/183 (9%)
Query: 258 FKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKS 317
F T N MKW +TE +GQ ++ D +++ A++NG +RGH + W Q P WV S
Sbjct: 41 FGQLTPENSMKWDATEPNRGQFTFSGSDYLVNFAQSNGKLIRGHTLVWH--SQLPGWVSS 98
Query: 318 LSPEE-LRKAAAKRINSVASRYSGELIAWDVVNE------NLHFNFFEGNLGENASAAYY 370
++ + L I +V +RY G++ AWDV+NE +L + F +GE+ +
Sbjct: 99 ITDKNTLISVLKNHITTVMTRYKGKIYAWDVLNEIFNEDGSLRNSVFYNVIGEDYVRIAF 158
Query: 371 STAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPL-AIGLQCHF 429
TA +DPN ++++N++N +++KK +AGIP+ IG Q H
Sbjct: 159 ETARSVDPNAKLYINDYNLDSAGYSKVNGMVSHVKKWL-------AAGIPIDGIGSQTHL 211
Query: 430 ASG 432
+G
Sbjct: 212 GAG 214
>XLNA_AGABI (O60206) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8)
(Xylanase)
Length = 333
Score = 92.8 bits (229), Expect = 2e-18
Identities = 64/209 (30%), Positives = 101/209 (47%), Gaps = 21/209 (10%)
Query: 236 FGCGINNNILTNSEY--QKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKN 293
FG +N L ++ Y Q + F T N MKW +TE +G ++ D + ++A+N
Sbjct: 31 FGTATDNPELGDAPYVAQLGNTADFNQITAGNSMKWDATEPSRGTFTFSNGDTVANMARN 90
Query: 294 NGISVRGHNIFWDDPKQQPQWVKS--LSPEELRKAAAKRINSVASRYSGELIAWDVVNE- 350
G +RGH W Q P WV S L +++ S Y G++ +WDVVNE
Sbjct: 91 RGQLLRGHTCVWH--SQLPNWVTSGNFDNSTLLSIVQNHCSTLVSHYRGQMYSWDVVNEP 148
Query: 351 -NLHFNFFEGNLGENASAAYYST----AHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIK 405
N +F + + AY +T A + DPNT++++N+FN IE +G + N ++
Sbjct: 149 FNEDGSFRQSVFFQKTGTAYIATALRAARNADPNTKLYINDFN-IEGTGAKSTGMINLVR 207
Query: 406 KIQQILQFPGSAGIPL-AIGLQCHFASGQ 433
+QQ +P+ IG+Q H GQ
Sbjct: 208 SLQQ-------QNVPIDGIGVQAHLIVGQ 229
>XYNA_ASPKA (P33559) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 327
Score = 92.4 bits (228), Expect = 3e-18
Identities = 59/202 (29%), Positives = 102/202 (50%), Gaps = 17/202 (8%)
Query: 239 GINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISV 298
G + NS+ + F T N MKW +TE +GQ +++ D +++ A++N +
Sbjct: 47 GDQYTLTKNSKTPAVIKADFGALTPENSMKWDATEPSRGQFSFSGSDYLVNFAQSNNKLI 106
Query: 299 RGHNIFWDDPKQQPQWVKSLSPEE-LRKAAAKRINSVASRYSGELIAWDVVNE------N 351
RGH + W Q P WV++++ + L + I +V Y G++ AWDVVNE +
Sbjct: 107 RGHTLVWH--SQLPSWVQAITDKNTLIEVMKNHITTVMQHYKGKIYAWDVVNEIFNEDGS 164
Query: 352 LHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQQIL 411
L + F +G++ + TA DPN ++++N++N S A +++KK +
Sbjct: 165 LRDSVFYKVIGDDYVRIAFETARAADPNAKLYINDYNLDSASYPKLAGMVSHVKKWIE-- 222
Query: 412 QFPGSAGIPL-AIGLQCHFASG 432
AGIP+ IG Q H ++G
Sbjct: 223 -----AGIPIDGIGSQTHLSAG 239
>XYNA_THESA (P36917) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 1157
Score = 90.1 bits (222), Expect = 1e-17
Identities = 102/449 (22%), Positives = 184/449 (40%), Gaps = 77/449 (17%)
Query: 51 GGIIVNPGFDH-NLEGWTVFGNGAIEERR--SNEGNAFIVARNRTQPLDSFSQKI--QLK 105
G +I N F++ N GW G+ ++ ++ G+ ++ RT + S + ++
Sbjct: 195 GNVIANETFENGNTSGWIGTGSSVVKAVYGVAHSGDYSLLTTGRTANWNGPSYDLTGKIV 254
Query: 106 KGMLYTFSAWLQLSEGSDTVSVVFKINGRE-----LIRGGHVIAKHGCWTLMKGGL---V 157
G Y W++ G+DT + + + G WT +KG V
Sbjct: 255 PGQQYNVDFWVKFVNGNDTEQIKATVKATSDKDNYIQVNDFANVNKGEWTEIKGSFTLPV 314
Query: 158 ANFTSPAEILFESKNPTVEIWADSVSLQPFTKEQWRSHQEESIERVRKSKVRFQVTHINE 217
A++ S I ES+NPT+E + D S+ I + +++ Q
Sbjct: 315 ADY-SGISIYVESQNPTLEFYIDDFSV---------------IGEISNNQITIQ------ 352
Query: 218 TALEGATITTKQTKLDFPFGCGINNNILTNSE-YQKWFLSRFKYTTFTNQMKWYSTEIVK 276
K FP G ++ + L +++ + + F N MK S + +
Sbjct: 353 ---NDIPDLYSVFKDYFPIGVAVDPSRLNDADPHAQLTAKHFNMLVAENAMKPESLQPTE 409
Query: 277 GQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWV--------KSLSPEELRKAAA 328
G + D ++ A + + +RGH + W + Q P W KS S + L +
Sbjct: 410 GNFTFDNADKIVDYAIAHNMKMRGHTLLWHN--QVPDWFFQDPSDPSKSASRDLLLQRLK 467
Query: 329 KRINSVASRY------SGELIAWDVVNENLHFNFFEGNL---------GENASAAYYSTA 373
I +V + +I WDVVNE L N GNL G + + A
Sbjct: 468 THITTVLDHFKTKYGSQNPIIGWDVVNEVLDDN---GNLRNSKWLQIIGPDYIEKAFEYA 524
Query: 374 HHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPL-AIGLQCHFASG 432
H DP+ ++F+N++N IE +G + + +KK++ S G+P+ IG+Q H +
Sbjct: 525 HEADPSMKLFINDYN-IENNGVKTQAMYDLVKKLK-------SEGVPIDGIGMQMHI-NI 575
Query: 433 QPNLAYMRAGLDLLGATGFPIWLTETSID 461
N+ ++A ++ L + G I +TE ++
Sbjct: 576 NSNIDNIKASIEKLASLGVEIQVTELDMN 604
>XYNA_PENCH (P29417) Endo-1,4-beta-xylanase precursor (EC 3.2.1.8)
(Xylanase) (1,4-beta-D-xylan xylanohydrolase)
Length = 353
Score = 90.1 bits (222), Expect = 1e-17
Identities = 55/176 (31%), Positives = 86/176 (48%), Gaps = 17/176 (9%)
Query: 265 NQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPE-EL 323
N MKW +TE +GQ ++ D + A+ NG +RGH + W Q P WV S++ + L
Sbjct: 77 NSMKWDATEPSQGQFSFAGSDYFVEFAETNGKLIRGHTLVWH--SQLPSWVSSITDKTTL 134
Query: 324 RKAAAKRINSVASRYSGELIAWDVVNE------NLHFNFFEGNLGENASAAYYSTAHHLD 377
I +V +Y G+L AWDVVNE L + F LGE+ + TA D
Sbjct: 135 TDVMKNHITTVMKQYKGKLYAWDVVNEIFEEDGTLRDSVFSRVLGEDFVRIAFETAREAD 194
Query: 378 PNTRMFLNEFNTIEYSGDVAASPTNYIKKIQQILQFPGSAGIPL-AIGLQCHFASG 432
P ++++N++N + +++KK +AG+P+ IG Q H +G
Sbjct: 195 PEAKLYINDYNLDSATSAKLQGMVSHVKKWI-------AAGVPIDGIGSQTHLGAG 243
>GUNB_CALSA (P10474) Endoglucanase/exoglucanase B precursor
[Includes: Endoglucanase (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Cellulase)
(Cellobiohydrolase); Exoglucanase (EC 3.2.1.91)
(Exocellobiohydrolase) (1,4-beta-cellobiohydrolase)]
Length = 1039
Score = 90.1 bits (222), Expect = 1e-17
Identities = 75/250 (30%), Positives = 112/250 (44%), Gaps = 37/250 (14%)
Query: 231 KLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQEN------YTTP 284
K DF G I L+N ++ L F T N+MK S ++ GQ + ++T
Sbjct: 50 KDDFMIGVAIPARCLSNDTDKRMVLKHFNSITAENEMKPES--LLAGQTSTGLSYRFSTA 107
Query: 285 DAMMSLAKNNGISVRGHNIFWDDPKQQPQWV------KSLSPEELRKAAAKRINSVASRY 338
DA + A N I +RGH + W + Q P W + LS + L + I V RY
Sbjct: 108 DAFVDFASTNKIGIRGHTLVWHN--QTPDWFFKDSNGQRLSKDALLARLKQYIYDVVGRY 165
Query: 339 SGELIAWDVVNENLHFN----FFEGNLGENASAAY----YSTAHHLDPNTRMFLNEFNT- 389
G++ AWDVVNE + N + E Y + AH DPN ++F N++NT
Sbjct: 166 KGKVYAWDVVNEAIDENQPDSYRRSTWYEICGPEYIEKAFIWAHEADPNAKLFYNDYNTE 225
Query: 390 IEYSGDVAASPTNYIKKIQQILQFPGSAGIPL-AIGLQCHFASGQPNLAYMRAGLDLLGA 448
I D N +K ++ S GIP+ IG+QCH P+++ + + L +
Sbjct: 226 ISKKRDFI---YNMVKNLK-------SKGIPIHGIGMQCHINVNWPSVSEIENSIKLFSS 275
Query: 449 -TGFPIWLTE 457
G I +TE
Sbjct: 276 IPGIEIHITE 285
>XYNA_PSEFL (P14768) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A) (XYLA)
Length = 611
Score = 86.7 bits (213), Expect = 2e-16
Identities = 69/254 (27%), Positives = 116/254 (45%), Gaps = 37/254 (14%)
Query: 233 DFPFGCGI-----NNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQE-NYTTPDA 286
DFP G + N +I T+S Q + F T N MK + + G ++T D
Sbjct: 271 DFPIGVAVAASGGNADIFTSSARQNIVRAEFNQITAENIMKM--SYMYSGSNFSFTNSDR 328
Query: 287 MMSLAKNNGISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRINSVASRYSGELIAWD 346
++S A NG +V GH + W Q P W S S R+ A+ I++VA+ ++G++ +WD
Sbjct: 329 LVSWAAQNGQTVHGHALVWHPSYQLPNWA-SDSNANFRQDFARHIDTVAAHFAGQVKSWD 387
Query: 347 VVNENLHFNFFEGNLGENASAAY------------------YSTAHHLDPNTRMFLNEFN 388
VVNE L F+ + G ++ Y + A DP ++ N+FN
Sbjct: 388 VVNEAL-FDSADDPDGRGSANGYRQSVFYRQFGGPEYIDEAFRRARAADPTAELYYNDFN 446
Query: 389 TIEYSGDVAASPTNYIKKIQQILQFPGSAGIPL-AIGLQCHFASGQPNLAYMRAGLDLLG 447
T E A T + +Q++L + G+P+ +G Q H + P++A +R + +
Sbjct: 447 TEEN----GAKTTALVNLVQRLL----NNGVPIDGVGFQMHVMNDYPSIANIRQAMQKIV 498
Query: 448 ATGFPIWLTETSID 461
A + + T +D
Sbjct: 499 ALSPTLKIKITELD 512
>GUNF_FUSOX (P46239) Putative endoglucanase type F precursor (EC
3.2.1.4) (Endo-1,4-beta-glucanase) (Cellulase)
Length = 385
Score = 86.3 bits (212), Expect = 2e-16
Identities = 56/205 (27%), Positives = 97/205 (47%), Gaps = 17/205 (8%)
Query: 236 FGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNG 295
FG I++ L N+ ++F T N MKW + E + ++ D ++ A NG
Sbjct: 96 FGTEIDHYHLNNNPLINIVKAQFGQVTCENSMKWDAIEPSRNSFTFSNADKVVDFATQNG 155
Query: 296 ISVRGHNIFWDDPKQQPQWVKSLSPEE-LRKAAAKRINSVASRYSGELIAWDVVNE---- 350
+RGH + W Q PQWV++++ L + ++ +RY G+++ WDVVN
Sbjct: 156 KLIRGHTLLWH--SQLPQWVQNINDRSTLTAVIENHVKTMVTRYKGKILQWDVVNNEIFA 213
Query: 351 ---NLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKI 407
NL + F LGE+ + A DP ++++N++N + D A + +
Sbjct: 214 EDGNLRDSVFSRVLGEDFVGIAFRAARAADPAAKLYINDYNLDK--SDYAKVTRGMVAHV 271
Query: 408 QQILQFPGSAGIPL-AIGLQCHFAS 431
+ + +AGIP+ IG Q H A+
Sbjct: 272 NKWI----AAGIPIDGIGSQGHLAA 292
>GUX_CELFI (P07986) Exoglucanase/xylanase precursor [Includes:
Exoglucanase (EC 3.2.1.91) (Exocellobiohydrolase)
(1,4-beta-cellobiohydrolase) (Beta-1,4-glycanase CEX);
Endo-1,4-beta-xylanase B (EC 3.2.1.8) (Xylanase B)]
Length = 484
Score = 84.0 bits (206), Expect = 1e-15
Identities = 61/233 (26%), Positives = 99/233 (42%), Gaps = 19/233 (8%)
Query: 236 FGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNG 295
FG ++ N L+ ++Y+ S F N MKW +TE + ++ D + S A + G
Sbjct: 56 FGFALDPNRLSEAQYKAIADSEFNLVVAENAMKWDATEPSQNSFSFGAGDRVASYAADTG 115
Query: 296 ISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRINSVASRYSGELIAWDVVNENL--- 352
+ GH + W Q P W K+L+ A + VA + G++ +WDVVNE
Sbjct: 116 KELYGHTLVWH--SQLPDWAKNLNGSAFESAMVNHVTKVADHFEGKVASWDVVNEAFADG 173
Query: 353 ----HFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIKKIQ 408
+ F+ LG + A DP ++ +N++N G A S + Y
Sbjct: 174 DGPPQDSAFQQKLGNGYIETAFRAARAADPTAKLCINDYNV---EGINAKSNSLY----- 225
Query: 409 QILQFPGSAGIPL-AIGLQCHFASGQPNLAYMRAGLDLLGATGFPIWLTETSI 460
+++ + G+PL +G Q H GQ R L G + +TE I
Sbjct: 226 DLVKDFKARGVPLDCVGFQSHLIVGQVP-GDFRQNLQRFADLGVDVRITELDI 277
>XYNA_DICTH (Q12603) Beta-1,4-xylanase (EC 3.2.1.8)
(Endo-1,4-beta-xylanase) (1,4-beta-D-xylan
xylanohydrolase)
Length = 352
Score = 82.4 bits (202), Expect = 3e-15
Identities = 61/236 (25%), Positives = 99/236 (41%), Gaps = 22/236 (9%)
Query: 234 FPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKN 293
F G +++ L Y+ F T NQMKW ++ D ++ A
Sbjct: 43 FTIGAAVSH--LNIYHYENLLKKHFNSLTPENQMKWEVIHPKPYVYDFGPADEIVDFAMK 100
Query: 294 NGISVRGHNIFWDDPKQQPQWVKSLSPEELRKAAAKRINSVASRYSGELIAWDVVNENLH 353
NG+ VRGH + W + Q P WV + + +E+ + I V Y G++ AWDVVNE L
Sbjct: 101 NGMKVRGHTLVWHN--QTPGWVYAGTKDEILARLKEHIKEVVGHYKGKVYAWDVVNEALS 158
Query: 354 FNFFE--------GNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIK 405
N E GE + AH +DP+ ++F N++N P +
Sbjct: 159 DNPNEFLRRAPWYDICGEEVIEKAFIWAHEVDPDAKLFYNDYN--------LEDPIKR-E 209
Query: 406 KIQQILQFPGSAGIPL-AIGLQCHFASGQPNLAYMRAGLDLLGATGFPIWLTETSI 460
K ++++ G+P+ IG+Q H+ P + + G + +TE I
Sbjct: 210 KAYKLVKKLKDKGVPIHGIGIQGHWTLAWPTPKMLEDSIKRFAELGVEVQVTEFDI 265
>XYNA_CALSR (P40944) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 684
Score = 80.5 bits (197), Expect = 1e-14
Identities = 67/257 (26%), Positives = 107/257 (41%), Gaps = 33/257 (12%)
Query: 234 FPFGCGINNNILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKN 293
F G + LTN + F T N+MK S + +G +++ D + K
Sbjct: 364 FKIGVAVPYRALTNPVDVEVIKRHFNSITPENEMKPESLQPYEGGFSFSIADEYVDFCKK 423
Query: 294 NGISVRGHNIFWDDPKQQPQWV------------KSLSPEELRKAAAKRINSVASRYSGE 341
+ IS+RGH + W +Q P W E L K I +V RY G+
Sbjct: 424 DNISLRGHTLVWH--QQTPSWFFTNPETGEKLTNSEKDKEILLDRLKKHIQTVVGRYKGK 481
Query: 342 LIAWDVVNENLHFNFFEGN--------LGENASAAYYSTAHHLDPNTRMFLNEFNTIEYS 393
+ AWDVVNE + N +G LG + AH DP ++F N+++T +
Sbjct: 482 VYAWDVVNEAIDENQPDGYRRSDWYNILGPEYIEKAFIWAHEADPKAKLFYNDYSTED-- 539
Query: 394 GDVAASPTNYIKKIQQILQFPGSAGIPL-AIGLQCHFASGQPNLAYMRAGLDLLG-ATGF 451
+I K+ + L+ + G+P+ +GLQCH + P+++ + + L G
Sbjct: 540 ----PYKREFIYKLIKNLK---AKGVPVHGVGLQCHISLDWPDVSEIEETVKLFSRIPGL 592
Query: 452 PIWLTETSIDPQPHQED 468
I TE I + D
Sbjct: 593 EIHFTEIDISIAKNMTD 609
>XYNZ_CLOTM (P10478) Endo-1,4-beta-xylanase Z precursor (EC 3.2.1.8)
(Xylanase Z) (1,4-beta-D-xylan xylanohydrolase Z)
Length = 837
Score = 80.1 bits (196), Expect = 1e-14
Identities = 52/208 (25%), Positives = 98/208 (47%), Gaps = 22/208 (10%)
Query: 237 GCGINNNILTNSE--YQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNN 294
G +N NS+ Y F N+MK+ + + + +++ D +++ A+ N
Sbjct: 530 GTCVNYPFYNNSDPTYNSILQREFSMVVCENEMKFDALQPRQNVFDFSKGDQLLAFAERN 589
Query: 295 GISVRGHNIFWDDPKQQPQWVK--SLSPEELRKAAAKRINSVASRYSGELIAWDVVNE-- 350
G+ +RGH + W + Q P W+ + + + L I +V + Y G+++ WDV NE
Sbjct: 590 GMQMRGHTLIWHN--QNPSWLTNGNWNRDSLLAVMKNHITTVMTHYKGKIVEWDVANECM 647
Query: 351 -----NLHFNFFEGNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGDVAASPTNYIK 405
L + + +G++ + A DP+ +F N++N IE G + + N IK
Sbjct: 648 DDSGNGLRSSIWRNVIGQDYLDYAFRYAREADPDALLFYNDYN-IEDLGPKSNAVFNMIK 706
Query: 406 KIQQILQFPGSAGIPL-AIGLQCHFASG 432
+++ G+P+ +G QCHF +G
Sbjct: 707 SMKE-------RGVPIDGVGFQCHFING 727
>XYNA_BACOV (P49942) Endo-1,4-beta-xylanase A precursor (EC 3.2.1.8)
(Xylanase A) (1,4-beta-D-xylan xylanohydrolase A)
Length = 376
Score = 73.2 bits (178), Expect = 2e-12
Identities = 64/274 (23%), Positives = 113/274 (40%), Gaps = 31/274 (11%)
Query: 208 VRFQVTHINETALEGATITTKQTKLDFPFGCGINNNILTNSEYQKWFLSRFKYTTFT--N 265
V F ++ A +G+++ K K F G +N + + + + + + + N
Sbjct: 13 VMFSFSYGEVFAKDGSSLK-KALKNKFLIGVSVNTHQSSGKDVAAVEIVKKNFNSIVAEN 71
Query: 266 QMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHNIFWDDPKQQPQWV------KSLS 319
MK + + N+ D +S ++N +++ GH + W Q W ++S
Sbjct: 72 CMKSSVIHPKENKYNFAQADEFVSFGESNQMAIIGHCLIWHS--QLAPWFCVDKDGNNVS 129
Query: 320 PEELRKAAAKRINSVASRYSGELIAWDVVNENLHFN-------FFEGNLGENASAAYYST 372
PE L+K I ++ RY G + WDVVNE + N F+E LGE +
Sbjct: 130 PEVLKKRMKDHITTIVKRYKGRIKGWDVVNEAIEDNGAYRKTKFYE-ILGEEYIPLAFQY 188
Query: 373 AHHLDPNTRMFLNEFNTIEYS-GDVAASPTNYIKKIQQILQFPGSAGIPL-AIGLQCHFA 430
AH DP+ ++ N+++ + + N +KK GI + AIG+Q H
Sbjct: 189 AHEADPDAELYYNDYSMAQPGRREAVVKMVNDLKK----------RGIRIDAIGMQGHIG 238
Query: 431 SGQPNLAYMRAGLDLLGATGFPIWLTETSIDPQP 464
P ++ + TG I +TE + P
Sbjct: 239 MDYPKISEFEKSMLAFAGTGVKIMITELDLTVIP 272
>XYNB_PSEFL (P23030) Endo-1,4-beta-xylanase B precursor (EC 3.2.1.8)
(Xylanase B) (1,4-beta-D-xylan xylanohydrolase B)
Length = 592
Score = 72.8 bits (177), Expect = 2e-12
Identities = 71/285 (24%), Positives = 110/285 (37%), Gaps = 29/285 (10%)
Query: 243 NILTNSEYQKWFLSRFKYTTFTNQMKWYSTEIVKGQENYTTPDAMMSLAKNNGISVRGHN 302
NI T+ + F + T N+ KW S E + N+ D + + A+ N I V+ H
Sbjct: 327 NITTSGAVRSDFTRYWNQITPENESKWGSVEGTRNVYNWAPLDRIYAYARQNNIPVKAHT 386
Query: 303 IFWDDPKQQPQWVKSLSPEELRKAAAKRINSVASRYSGELIAWDVVNENLHFN----FFE 358
W Q P W+ +LS E+ + I +RY + DVVNE + + + +
Sbjct: 387 FVWG--AQSPSWLNNLSGPEVAVEIEQWIRDYCARYPDTAMI-DVVNEAVPGHQPAGYAQ 443
Query: 359 GNLGENASAAYYSTAHHLDPNTRMFLNEFNTIEYSGD----VAASPTNYIKKIQQILQFP 414
G N + A PN+ + LN++N I + + +A + NYI
Sbjct: 444 RAFGNNWIQRVFQLARQYCPNSILILNDYNNIRWQHNEFIALAKAQGNYID--------- 494
Query: 415 GSAGIPLAIGLQCHFASGQPNLAYMRAGLDLLGATGFPIWLTETSIDPQPHQEDY--FDW 472
A+GLQ H G A ++ G PI+++E I Q F
Sbjct: 495 -------AVGLQAHELKGMTAAQVKTAIDNIWNQVGKPIYISEYDIGDTNDQVQLQNFQA 547
Query: 473 ILREGYAHPAVQGIIMFMGPVQAGFKKAPLADENFKNTPIGDVVD 517
Y HP V GI + Q ++ + P DVVD
Sbjct: 548 HFPVFYNHPHVHGITSGICGGQDLDRRLRFDPGQWHTAPGNDVVD 592
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 72,386,958
Number of Sequences: 164201
Number of extensions: 3228240
Number of successful extensions: 7206
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 7109
Number of HSP's gapped (non-prelim): 53
length of query: 581
length of database: 59,974,054
effective HSP length: 116
effective length of query: 465
effective length of database: 40,926,738
effective search space: 19030933170
effective search space used: 19030933170
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0218.17


