

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0215b.7
(68 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
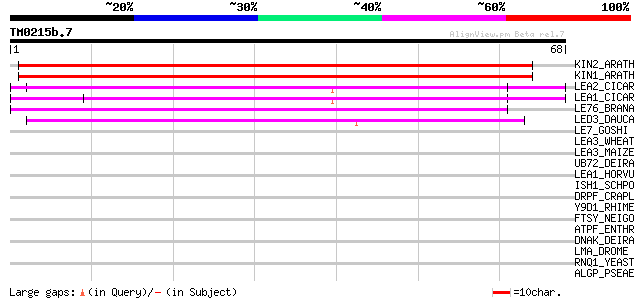
Score E
Sequences producing significant alignments: (bits) Value
KIN2_ARATH (P31169) Stress-induced KIN2 protein (Cold-induced CO... 55 3e-08
KIN1_ARATH (P18612) Stress-induced KIN1 protein 52 3e-07
LEA2_CICAR (O49817) Late embryogenesis abundant protein 2 (CapLE... 50 1e-06
LEA1_CICAR (O49816) Late embryogenesis abundant protein 1 (CapLE... 45 3e-05
LE76_BRANA (P13934) Late embryogenesis abundant protein 76 (LEA 76) 42 3e-04
LED3_DAUCA (P83442) Late embryogenesis abundant protein Dc3 40 9e-04
LE7_GOSHI (P13939) Late embryogenesis abundant protein D-7 (LEA ... 40 0.001
LEA3_WHEAT (Q03968) Late embryogenesis abundant protein, group 3... 39 0.002
LEA3_MAIZE (Q42376) Late embryogenesis abundant protein, group 3... 38 0.006
UB72_DEIRA (Q9RV58) Protein DR1172 36 0.016
LEA1_HORVU (P14928) ABA-inducible protein PHV A1 36 0.016
ISH1_SCHPO (Q9Y7X6) Stress response protein ish1 33 0.14
DRPF_CRAPL (P23283) Desiccation-related protein PCC3-06 33 0.18
Y9D1_RHIME (Q92XV9) Hypothetical UPF0337 protein RA1131 32 0.30
FTSY_NEIGO (P14929) Cell division protein ftsY homolog 32 0.40
ATPF_ENTHR (P26681) ATP synthase B chain (EC 3.6.3.14) 31 0.68
DNAK_DEIRA (Q9RY23) Chaperone protein dnaK (Heat shock protein 7... 30 0.88
LMA_DROME (Q00174) Laminin alpha chain precursor 29 2.0
RNQ1_YEAST (P25367) [PIN+] prion protein RNQ1 (Rich in asparagin... 29 2.6
ALGP_PSEAE (P15276) Transcriptional regulatory protein algP (Alg... 29 2.6
>KIN2_ARATH (P31169) Stress-induced KIN2 protein (Cold-induced
COR6.6 protein)
Length = 66
Score = 55.1 bits (131), Expect = 3e-08
Identities = 27/63 (42%), Positives = 43/63 (67%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
++ K ++ AGQA G+AEEK++ L+D A +AA +A + Q AG+ I +A G + VK+ T
Sbjct: 3 ETNKNAFQAGQAAGKAEEKSNVLLDKAKDAAAAAGASAQQAGKSISDAAVGGVNFVKDKT 62
Query: 62 GMN 64
G+N
Sbjct: 63 GLN 65
>KIN1_ARATH (P18612) Stress-induced KIN1 protein
Length = 66
Score = 52.0 bits (123), Expect = 3e-07
Identities = 25/63 (39%), Positives = 40/63 (62%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
++ K ++ AGQ G+AEEK++ L+D A +AA A Q AG+ + +A G + VK+ T
Sbjct: 3 ETNKNAFQAGQTAGKAEEKSNVLLDKAKDAAAGAGAGAQQAGKSVSDAAAGGVNFVKDKT 62
Query: 62 GMN 64
G+N
Sbjct: 63 GLN 65
>LEA2_CICAR (O49817) Late embryogenesis abundant protein 2
(CapLEA-2)
Length = 155
Score = 49.7 bits (117), Expect = 1e-06
Identities = 26/61 (42%), Positives = 33/61 (53%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M S SY AG+AKG+ EEKT+ ++ + AQ+AKE Q A Q K A A K
Sbjct: 1 MTSHDQSYRAGEAKGRTEEKTNQMIGNIEDKAQAAKEKAQQAAQTAKDKTSQTAQAAKEK 60
Query: 61 T 61
T
Sbjct: 61 T 61
Score = 43.5 bits (101), Expect = 1e-04
Identities = 25/70 (35%), Positives = 35/70 (49%), Gaps = 4/70 (5%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKET----VQGAGQQIKASAQGAADAVK 58
+Q A G+A+EK S + AQS K+ +Q G++ K AQGA DAVK
Sbjct: 76 TQATKEKAQDTTGRAKEKGSEMGQSTKETAQSGKDNSAGFLQQTGEKAKGMAQGATDAVK 135
Query: 59 NATGMNNNSK 68
GM N+ +
Sbjct: 136 QTFGMTNDDQ 145
>LEA1_CICAR (O49816) Late embryogenesis abundant protein 1
(CapLEA-1)
Length = 177
Score = 45.4 bits (106), Expect = 3e-05
Identities = 24/61 (39%), Positives = 31/61 (50%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M S SY AG+ G+ EEKT+ ++ + AQ+AKE Q A Q K A A K
Sbjct: 1 MASHDQSYKAGETMGRTEEKTNQMIGNIEDKAQAAKEKAQQAAQTAKDKTSQTAQAAKEK 60
Query: 61 T 61
T
Sbjct: 61 T 61
Score = 44.7 bits (104), Expect = 5e-05
Identities = 25/63 (39%), Positives = 33/63 (51%), Gaps = 4/63 (6%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKET----VQGAGQQIKASAQGAADAVKNATGMNN 65
A G+A EK S + AQS K+ +Q G+++K AQGA DAVK GM N
Sbjct: 105 AQDTTGRAREKGSEMGQSTKETAQSGKDNSAGFLQQTGEKVKGMAQGATDAVKQTFGMAN 164
Query: 66 NSK 68
+ K
Sbjct: 165 DDK 167
>LE76_BRANA (P13934) Late embryogenesis abundant protein 76 (LEA
76)
Length = 280
Score = 42.0 bits (97), Expect = 3e-04
Identities = 20/61 (32%), Positives = 31/61 (50%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M S + SY AG+ +G+ +EKT M + A+ K+ Q+ + AQ A A K+
Sbjct: 1 MASNQQSYKAGETRGKTQEKTGQAMGAMRDKAEEGKDKTSQTAQKAQQKAQETAQAAKDK 60
Query: 61 T 61
T
Sbjct: 61 T 61
Score = 37.7 bits (86), Expect = 0.006
Identities = 23/69 (33%), Positives = 32/69 (46%), Gaps = 11/69 (15%)
Query: 6 MSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGA-----------GQQIKASAQGAA 54
+S K +A++ + A NAAQ KET + G+ +K A GAA
Sbjct: 149 LSETGEAVKQKAQDAAQYTKETAQNAAQYTKETAEAGKDKTGGFLSQTGEHVKQMAMGAA 208
Query: 55 DAVKNATGM 63
DAVK+ GM
Sbjct: 209 DAVKHTFGM 217
Score = 30.4 bits (67), Expect = 0.88
Identities = 20/63 (31%), Positives = 28/63 (43%), Gaps = 4/63 (6%)
Query: 3 SQKMSYNAGQAKGQAEEKTS----NLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVK 58
+QK A + A++KTS A AQ+AK+ A Q + AQ A A K
Sbjct: 43 AQKAQQKAQETAQAAKDKTSQAAQTTQQKAQETAQAAKDKTSQAAQTTQQKAQETAQAAK 102
Query: 59 NAT 61
+ T
Sbjct: 103 DKT 105
>LED3_DAUCA (P83442) Late embryogenesis abundant protein Dc3
Length = 163
Score = 40.4 bits (93), Expect = 9e-04
Identities = 24/65 (36%), Positives = 32/65 (48%), Gaps = 4/65 (6%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQG----AGQQIKASAQGAADAVK 58
+Q G A +EK S + + A A + KE G A +Q+K AQGA +AVK
Sbjct: 77 AQAAKEKTGGAMQATKEKASEMGESAKETAVAGKEKTGGLMSSAAEQVKGMAQGATEAVK 136
Query: 59 NATGM 63
N GM
Sbjct: 137 NTFGM 141
Score = 38.1 bits (87), Expect = 0.004
Identities = 21/59 (35%), Positives = 31/59 (51%), Gaps = 4/59 (6%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNATG 62
Q SY AG+ KG A+EKT + D + AQ+AK+ ++ SA+ K+ TG
Sbjct: 5 QDQSYKAGEPKGHAQEKTGQMADTMKDKAQAAKD----KASEMAGSARDRTVESKDQTG 59
>LE7_GOSHI (P13939) Late embryogenesis abundant protein D-7 (LEA
D-7)
Length = 136
Score = 39.7 bits (91), Expect = 0.001
Identities = 22/70 (31%), Positives = 36/70 (51%), Gaps = 4/70 (5%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAK----ETVQGAGQQIKASAQGAADA 56
M S + SY AG+A+G+A EK + + A++AK ET + A Q+ +A+ A
Sbjct: 1 MASHEQSYKAGRAEGRAHEKGEQMKESMKEKAEAAKQKTMETAEAAKQKTMETAEAAKQK 60
Query: 57 VKNATGMNNN 66
+ A N+
Sbjct: 61 TRGAAETTND 70
Score = 38.5 bits (88), Expect = 0.003
Identities = 23/59 (38%), Positives = 34/59 (56%), Gaps = 7/59 (11%)
Query: 12 QAKGQAE---EKTSNLMDMASNAAQSAKET----VQGAGQQIKASAQGAADAVKNATGM 63
+ +G AE +KT A A+ KET +Q AG++++ +AQGA DAVK+ GM
Sbjct: 60 KTRGAAETTNDKTKQTAGAARGKAEETKETSGGILQQAGEKVRNAAQGATDAVKHTFGM 118
>LEA3_WHEAT (Q03968) Late embryogenesis abundant protein, group 3
(LEA) (PMA2005)
Length = 224
Score = 38.9 bits (89), Expect = 0.002
Identities = 21/60 (35%), Positives = 35/60 (58%), Gaps = 1/60 (1%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASN-AAQSAKETVQGAGQQIKASAQGAADAVKNA 60
+ + SY+AG+ K + EEKT +M + A Q+ + T Q AG+ +A+ Q AA+ + A
Sbjct: 4 NQNQASYHAGETKARNEEKTGQVMGATKDKAGQTTEATKQKAGETTEATKQKAAETTEAA 63
Score = 33.1 bits (74), Expect = 0.14
Identities = 23/75 (30%), Positives = 33/75 (43%), Gaps = 11/75 (14%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKET-----------VQGAGQQIKASAQG 52
QK + A+ +A E T AS AQ KE+ +Q AG+ + + G
Sbjct: 127 QKAAETTEAARQKAAEATEAAKQKASETAQYTKESAVTGKDKTGSVLQQAGETVVNAVVG 186
Query: 53 AADAVKNATGMNNNS 67
A DAV N GM ++
Sbjct: 187 AKDAVANTLGMGGDN 201
>LEA3_MAIZE (Q42376) Late embryogenesis abundant protein, group 3
(LEA)
Length = 221
Score = 37.7 bits (86), Expect = 0.006
Identities = 19/54 (35%), Positives = 25/54 (46%)
Query: 5 KMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVK 58
K SY AG+ K + EEKT + + AQ AK+ A Q A +A K
Sbjct: 7 KASYQAGETKARTEEKTGQAVGATKDTAQHAKDRAADAAGHAAGKGQDAKEATK 60
Score = 35.4 bits (80), Expect = 0.027
Identities = 23/74 (31%), Positives = 38/74 (51%), Gaps = 15/74 (20%)
Query: 10 AGQAKGQAEEKTSNLMDMA----SNAAQSAKET-----------VQGAGQQIKASAQGAA 54
AG+ A +K ++ M+ A + A Q AK+T +Q A +Q+K++A G
Sbjct: 125 AGETTEAARQKAADAMEAAKQKAAEAGQYAKDTAVSGKDKSGGVIQQATEQVKSAAAGRK 184
Query: 55 DAVKNATGMNNNSK 68
DAV + GM ++K
Sbjct: 185 DAVMSTLGMGGDNK 198
>UB72_DEIRA (Q9RV58) Protein DR1172
Length = 298
Score = 36.2 bits (82), Expect = 0.016
Identities = 19/51 (37%), Positives = 27/51 (52%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGA 53
+Q + A QA A++K ++ AS AA AK+ Q Q +K SAQ A
Sbjct: 196 AQNVKQGAQQAASDAKDKVQDVKADASRAADQAKDKAQDVAQNVKQSAQDA 246
Score = 33.9 bits (76), Expect = 0.080
Identities = 16/50 (32%), Positives = 23/50 (46%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKN 59
AG+ K T+ + D A +AK+ Q GQ +K A AD K+
Sbjct: 83 AGEVKSAVAGATAEIKDAGKEVADTAKDAGQNVGQNVKREAADLADQAKD 132
Score = 33.5 bits (75), Expect = 0.10
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 4/50 (8%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKN 59
A QAK +A++ N+ A AA AK+ V Q +KA A AAD K+
Sbjct: 185 ADQAKDKAQDVAQNVKQGAQQAASDAKDKV----QDVKADASRAADQAKD 230
Score = 32.0 bits (71), Expect = 0.30
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 4/54 (7%)
Query: 6 MSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKN 59
+S A QAK +A++ N+ A AA + K+ VQ +KA A AAD K+
Sbjct: 141 VSKAADQAKDKAQDVAQNVQAGAQQAAANVKDKVQ----DVKADASKAADQAKD 190
>LEA1_HORVU (P14928) ABA-inducible protein PHV A1
Length = 213
Score = 36.2 bits (82), Expect = 0.016
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query: 7 SYNAGQAKGQAEEKTSNLMDMA-SNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
SY+AG+ K + EEKT +M A Q+ + T Q AG+ +A+ Q + + A
Sbjct: 9 SYHAGETKARTEEKTGQMMGATKQKAGQTTEATKQKAGETAEATKQKTGETAEAA 63
Score = 34.7 bits (78), Expect = 0.047
Identities = 24/75 (32%), Positives = 34/75 (45%), Gaps = 11/75 (14%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKET-----------VQGAGQQIKASAQG 52
QK + AK +A E T AS+ AQ KE+ +Q AG+ + + G
Sbjct: 116 QKAAETTEAAKQKAAEATEAAKQKASDTAQYTKESAVAGKDKTGSVLQQAGETVVNAVVG 175
Query: 53 AADAVKNATGMNNNS 67
A DAV N GM ++
Sbjct: 176 AKDAVANTLGMGGDN 190
>ISH1_SCHPO (Q9Y7X6) Stress response protein ish1
Length = 684
Score = 33.1 bits (74), Expect = 0.14
Identities = 20/59 (33%), Positives = 29/59 (48%), Gaps = 4/59 (6%)
Query: 14 KGQAEEKTSNLMD----MASNAAQSAKETVQGAGQQIKASAQGAADAVKNATGMNNNSK 68
K +A E+ SN+ D +ASN A A E QGA +GA A ++ T ++K
Sbjct: 344 KSKAYEEASNVADSASSIASNVASGATEAYQGAASGASRFTEGAKTAAESVTSAFEHNK 402
>DRPF_CRAPL (P23283) Desiccation-related protein PCC3-06
Length = 201
Score = 32.7 bits (73), Expect = 0.18
Identities = 23/83 (27%), Positives = 34/83 (40%), Gaps = 27/83 (32%)
Query: 2 DSQKMSYNA-----GQAKGQAEEKTSNLMDMA----------------------SNAAQS 34
D K +YNA G+ K + +E N+ + A SN Q+
Sbjct: 94 DKAKETYNAASGKAGELKDKTQEGAENVREKAMDAGNDAMEKTRNAGERVADGVSNVGQN 153
Query: 35 AKETVQGAGQQIKASAQGAADAV 57
KE V GAG+++K A+ D V
Sbjct: 154 VKENVMGAGEKVKEFAEDVKDTV 176
>Y9D1_RHIME (Q92XV9) Hypothetical UPF0337 protein RA1131
Length = 63
Score = 32.0 bits (71), Expect = 0.30
Identities = 19/46 (41%), Positives = 25/46 (54%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAAD 55
AG+AK A + T N A AAQ AK Q A ++K + +GA D
Sbjct: 16 AGKAKKAAGDATDNNSLRAKGAAQEAKGGAQQAKGKLKDAVKGAVD 61
Score = 27.7 bits (60), Expect = 5.7
Identities = 18/54 (33%), Positives = 25/54 (45%), Gaps = 4/54 (7%)
Query: 11 GQAKGQAEEKTSNLMDMASNAAQSAKET----VQGAGQQIKASAQGAADAVKNA 60
G AK + K + L A AA A + +GA Q+ K AQ A +K+A
Sbjct: 2 GSAKDKVAGKANELAGKAKKAAGDATDNNSLRAKGAAQEAKGGAQQAKGKLKDA 55
>FTSY_NEIGO (P14929) Cell division protein ftsY homolog
Length = 416
Score = 31.6 bits (70), Expect = 0.40
Identities = 20/63 (31%), Positives = 31/63 (48%), Gaps = 7/63 (11%)
Query: 5 KMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIK-------ASAQGAADAV 57
K+ Q G +E +L + A+SA ETV GA +Q+K + A AA+ V
Sbjct: 27 KVESEVAQIVGNIKEDVESLAESVKGRAESAVETVSGAVEQVKETVAEMPSEAGEAAERV 86
Query: 58 KNA 60
++A
Sbjct: 87 ESA 89
>ATPF_ENTHR (P26681) ATP synthase B chain (EC 3.6.3.14)
Length = 174
Score = 30.8 bits (68), Expect = 0.68
Identities = 16/43 (37%), Positives = 25/43 (57%)
Query: 18 EEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
E+K +N +D A + ++ + Q QQ+ AS AAD +KNA
Sbjct: 48 EDKIANDLDSAEKSRINSAKMEQEREQQLLASRSDAADIIKNA 90
>DNAK_DEIRA (Q9RY23) Chaperone protein dnaK (Heat shock protein 70)
(Heat shock 70 kDa protein) (HSP70)
Length = 627
Score = 30.4 bits (67), Expect = 0.88
Identities = 22/67 (32%), Positives = 32/67 (46%), Gaps = 3/67 (4%)
Query: 4 QKMSYNAGQAKGQAE--EKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
Q+ NA K + E EK +NL + A Q +E +GA Q K + AAD + A
Sbjct: 512 QEAEQNAAADKQRKEKVEKRNNLDSLRVQAVQQLEEQ-EGAAQDAKDRLKAAADEAEEAV 570
Query: 62 GMNNNSK 68
++SK
Sbjct: 571 RSEDDSK 577
>LMA_DROME (Q00174) Laminin alpha chain precursor
Length = 3712
Score = 29.3 bits (64), Expect = 2.0
Identities = 17/70 (24%), Positives = 35/70 (49%), Gaps = 7/70 (10%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMA-------SNAAQSAKETVQGAGQQIKASAQGAAD 55
+QK+S +A A G A +KT + + A ++ Q A++++Q ++ +A
Sbjct: 2469 AQKLSQDAISAAGNATDKTDGIEERAHLADTGSTDLLQRARQSLQKVQDDLEPRLNASAG 2528
Query: 56 AVKNATGMNN 65
V+ + +NN
Sbjct: 2529 KVQKISAVNN 2538
>RNQ1_YEAST (P25367) [PIN+] prion protein RNQ1 (Rich in asparagine
and glutamine protein 1)
Length = 405
Score = 28.9 bits (63), Expect = 2.6
Identities = 17/65 (26%), Positives = 30/65 (46%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNATGM 63
Q GQ +GQ + + L +AS+ S QG Q S+ GA ++ ++
Sbjct: 157 QGQGQGQGQGQGQGQGSFTALASLASSFMNSNNNNQQGQNQSSGGSSFGALASMASSFMH 216
Query: 64 NNNSK 68
+NN++
Sbjct: 217 SNNNQ 221
>ALGP_PSEAE (P15276) Transcriptional regulatory protein algP
(Alginate regulatory protein algR3)
Length = 352
Score = 28.9 bits (63), Expect = 2.6
Identities = 14/58 (24%), Positives = 29/58 (49%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVK 58
+DS+K+ + +G+A+EK +AA++ K Q ++ + + A D +K
Sbjct: 35 VDSEKLLAKLEKQRGKAQEKLHKARTKLQDAAKAGKTKAQAKARETISDLEEALDTLK 92
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.295 0.108 0.262
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,662,144
Number of Sequences: 164201
Number of extensions: 136663
Number of successful extensions: 418
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 354
Number of HSP's gapped (non-prelim): 62
length of query: 68
length of database: 59,974,054
effective HSP length: 44
effective length of query: 24
effective length of database: 52,749,210
effective search space: 1265981040
effective search space used: 1265981040
T: 11
A: 40
X1: 17 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0215b.7


