

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
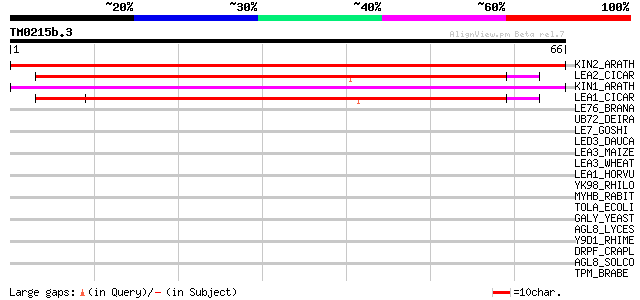
Query= TM0215b.3
(66 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
KIN2_ARATH (P31169) Stress-induced KIN2 protein (Cold-induced CO... 51 6e-07
LEA2_CICAR (O49817) Late embryogenesis abundant protein 2 (CapLE... 48 5e-06
KIN1_ARATH (P18612) Stress-induced KIN1 protein 48 5e-06
LEA1_CICAR (O49816) Late embryogenesis abundant protein 1 (CapLE... 46 2e-05
LE76_BRANA (P13934) Late embryogenesis abundant protein 76 (LEA 76) 40 0.001
UB72_DEIRA (Q9RV58) Protein DR1172 40 0.001
LE7_GOSHI (P13939) Late embryogenesis abundant protein D-7 (LEA ... 39 0.002
LED3_DAUCA (P83442) Late embryogenesis abundant protein Dc3 36 0.016
LEA3_MAIZE (Q42376) Late embryogenesis abundant protein, group 3... 36 0.021
LEA3_WHEAT (Q03968) Late embryogenesis abundant protein, group 3... 35 0.047
LEA1_HORVU (P14928) ABA-inducible protein PHV A1 33 0.14
YK98_RHILO (Q98J52) Hypothetical UPF0337 protein mlr2098 32 0.40
MYHB_RABIT (P35748) Myosin heavy chain, smooth muscle isoform (S... 32 0.40
TOLA_ECOLI (P19934) TolA protein 31 0.68
GALY_YEAST (P19659) Transcription regulatory protein GAL11 30 0.89
AGL8_LYCES (Q40170) Agamous-like MADS box protein AGL8 homolog (... 30 0.89
Y9D1_RHIME (Q92XV9) Hypothetical UPF0337 protein RA1131 29 2.0
DRPF_CRAPL (P23283) Desiccation-related protein PCC3-06 29 2.6
AGL8_SOLCO (O22328) Agamous-like MADS box protein AGL8 homolog 29 2.6
TPM_BRABE (Q9NDS0) Tropomyosin 28 3.4
>KIN2_ARATH (P31169) Stress-induced KIN2 protein (Cold-induced
COR6.6 protein)
Length = 66
Score = 50.8 bits (120), Expect = 6e-07
Identities = 25/66 (37%), Positives = 41/66 (61%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
M ++ + QAGQA G+A+EK++ ++ A++AA +A S Q+AG+ + A G + VK
Sbjct: 1 MSETNKNAFQAGQAAGKAEEKSNVLLDKAKDAAAAAGASAQQAGKSISDAAVGGVNFVKD 60
Query: 61 AIGANK 66
G NK
Sbjct: 61 KTGLNK 66
>LEA2_CICAR (O49817) Late embryogenesis abundant protein 2
(CapLEA-2)
Length = 155
Score = 47.8 bits (112), Expect = 5e-06
Identities = 22/56 (39%), Positives = 35/56 (62%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S + S++AG+A G+ +EK + M+GN + AQ+A++ Q+A Q K K A A K
Sbjct: 3 SHDQSYRAGEAKGRTEEKTNQMIGNIEDKAQAAKEKAQQAAQTAKDKTSQTAQAAK 58
Score = 43.9 bits (102), Expect = 8e-05
Identities = 24/64 (37%), Positives = 35/64 (54%), Gaps = 4/64 (6%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQS----CQEAGQQMKAKAQGAADAVK 59
+Q +A TG+A+EK S M + + AQS + + Q+ G++ K AQGA DAVK
Sbjct: 76 TQATKEKAQDTTGRAKEKGSEMGQSTKETAQSGKDNSAGFLQQTGEKAKGMAQGATDAVK 135
Query: 60 SAIG 63
G
Sbjct: 136 QTFG 139
>KIN1_ARATH (P18612) Stress-induced KIN1 protein
Length = 66
Score = 47.8 bits (112), Expect = 5e-06
Identities = 23/66 (34%), Positives = 38/66 (56%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
M ++ + QAGQ G+A+EK++ ++ A++AA A Q+AG+ + A G + VK
Sbjct: 1 MSETNKNAFQAGQTAGKAEEKSNVLLDKAKDAAAGAGAGAQQAGKSVSDAAAGGVNFVKD 60
Query: 61 AIGANK 66
G NK
Sbjct: 61 KTGLNK 66
>LEA1_CICAR (O49816) Late embryogenesis abundant protein 1
(CapLEA-1)
Length = 177
Score = 46.2 bits (108), Expect = 2e-05
Identities = 21/56 (37%), Positives = 34/56 (60%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S + S++AG+ G+ +EK + M+GN + AQ+A++ Q+A Q K K A A K
Sbjct: 3 SHDQSYKAGETMGRTEEKTNQMIGNIEDKAQAAKEKAQQAAQTAKDKTSQTAQAAK 58
Score = 43.5 bits (101), Expect = 1e-04
Identities = 23/58 (39%), Positives = 34/58 (57%), Gaps = 4/58 (6%)
Query: 10 QAGQATGQAQEKASNMMGNARNAAQSAQQSC----QEAGQQMKAKAQGAADAVKSAIG 63
+A TG+A+EK S M + + AQS + + Q+ G+++K AQGA DAVK G
Sbjct: 104 KAQDTTGRAREKGSEMGQSTKETAQSGKDNSAGFLQQTGEKVKGMAQGATDAVKQTFG 161
>LE76_BRANA (P13934) Late embryogenesis abundant protein 76 (LEA
76)
Length = 280
Score = 40.0 bits (92), Expect = 0.001
Identities = 19/56 (33%), Positives = 29/56 (50%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S S++AG+ G+ QEK MG R+ A+ + + Q+ + KAQ A A K
Sbjct: 3 SNQQSYKAGETRGKTQEKTGQAMGAMRDKAEEGKDKTSQTAQKAQQKAQETAQAAK 58
Score = 34.7 bits (78), Expect = 0.047
Identities = 23/74 (31%), Positives = 37/74 (49%), Gaps = 12/74 (16%)
Query: 2 DQSQNLSHQAGQATGQ-AQEKASNMMGNARNAAQSAQQSCQ-----------EAGQQMKA 49
D++ + + G+A Q AQ+ A A+NAAQ +++ + + G+ +K
Sbjct: 143 DKTGSYLSETGEAVKQKAQDAAQYTKETAQNAAQYTKETAEAGKDKTGGFLSQTGEHVKQ 202
Query: 50 KAQGAADAVKSAIG 63
A GAADAVK G
Sbjct: 203 MAMGAADAVKHTFG 216
Score = 33.1 bits (74), Expect = 0.14
Identities = 21/57 (36%), Positives = 27/57 (46%), Gaps = 1/57 (1%)
Query: 4 SQNLSHQAGQATGQ-AQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
+++ + QA Q T Q AQE A AAQ+ QQ QE Q K K AA +
Sbjct: 57 AKDKTSQAAQTTQQKAQETAQAAKDKTSQAAQTTQQKAQETAQAAKDKTSQAAQTTQ 113
Score = 32.0 bits (71), Expect = 0.31
Identities = 19/52 (36%), Positives = 23/52 (43%)
Query: 8 SHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S A +A +AQE A AAQ+ QQ QE Q K K AA +
Sbjct: 40 SQTAQKAQQKAQETAQAAKDKTSQAAQTTQQKAQETAQAAKDKTSQAAQTTQ 91
Score = 31.2 bits (69), Expect = 0.52
Identities = 20/57 (35%), Positives = 26/57 (45%), Gaps = 1/57 (1%)
Query: 4 SQNLSHQAGQATGQ-AQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
+++ + QA Q T Q AQE A AAQ+ QQ E Q K K AA +
Sbjct: 79 AKDKTSQAAQTTQQKAQETAQAAKDKTSQAAQTTQQKAHETTQSSKEKTSQAAQTAQ 135
Score = 27.7 bits (60), Expect = 5.8
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 12/68 (17%)
Query: 4 SQNLSHQAGQATGQ-AQEKASNMMGNARNAAQSAQQSCQ-----------EAGQQMKAKA 51
+++ + QA Q T Q A E + AAQ+AQ+ + E G+ +K KA
Sbjct: 101 AKDKTSQAAQTTQQKAHETTQSSKEKTSQAAQTAQEKARETKDKTGSYLSETGEAVKQKA 160
Query: 52 QGAADAVK 59
Q AA K
Sbjct: 161 QDAAQYTK 168
>UB72_DEIRA (Q9RV58) Protein DR1172
Length = 298
Score = 39.7 bits (91), Expect = 0.001
Identities = 20/58 (34%), Positives = 32/58 (54%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
D+ Q++ A +A QA++KA ++ N + AQ A ++ Q +KA A AAD K
Sbjct: 172 DKVQDVKADASKAADQAKDKAQDVAQNVKQGAQQAASDAKDKVQDVKADASRAADQAK 229
Score = 38.9 bits (89), Expect = 0.002
Identities = 19/58 (32%), Positives = 31/58 (52%)
Query: 5 QNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAI 62
QN+ +A QA++KA ++ + AA A+ Q+ Q ++A AQ AA VK +
Sbjct: 117 QNVKREAADLADQAKDKAQDVKADVSKAADQAKDKAQDVAQNVQAGAQQAAANVKDKV 174
Score = 37.7 bits (86), Expect = 0.006
Identities = 19/58 (32%), Positives = 33/58 (56%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
D++Q++ +A QA++KA ++ N + AQ A + ++ Q +KA A AAD K
Sbjct: 132 DKAQDVKADVSKAADQAKDKAQDVAQNVQAGAQQAAANVKDKVQDVKADASKAADQAK 189
Score = 32.0 bits (71), Expect = 0.31
Identities = 22/76 (28%), Positives = 37/76 (47%), Gaps = 11/76 (14%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAA-----------QSAQQSCQEAGQQMKAK 50
D +QN+ A QA A++K ++ +A AA Q+ +QS Q+A + AK
Sbjct: 194 DVAQNVKQGAQQAASDAKDKVQDVKADASRAADQAKDKAQDVAQNVKQSAQDAKTDVDAK 253
Query: 51 AQGAADAVKSAIGANK 66
A+ A +++ A K
Sbjct: 254 AKSWAFDLRTDAEAGK 269
>LE7_GOSHI (P13939) Late embryogenesis abundant protein D-7 (LEA
D-7)
Length = 136
Score = 39.3 bits (90), Expect = 0.002
Identities = 19/56 (33%), Positives = 30/56 (52%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S S++AG+A G+A EK M + + A++A+Q E + K K A+A K
Sbjct: 3 SHEQSYKAGRAEGRAHEKGEQMKESMKEKAEAAKQKTMETAEAAKQKTMETAEAAK 58
Score = 34.7 bits (78), Expect = 0.047
Identities = 19/67 (28%), Positives = 33/67 (48%), Gaps = 4/67 (5%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSC----QEAGQQMKAKAQGAAD 56
M+ ++ + A +K G AR A+ +++ Q+AG++++ AQGA D
Sbjct: 51 METAEAAKQKTRGAAETTNDKTKQTAGAARGKAEETKETSGGILQQAGEKVRNAAQGATD 110
Query: 57 AVKSAIG 63
AVK G
Sbjct: 111 AVKHTFG 117
>LED3_DAUCA (P83442) Late embryogenesis abundant protein Dc3
Length = 163
Score = 36.2 bits (82), Expect = 0.016
Identities = 21/66 (31%), Positives = 34/66 (50%), Gaps = 4/66 (6%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSC----QEAGQQMKAKAQGAADA 57
+ +Q + G A +EKAS M +A+ A + ++ A +Q+K AQGA +A
Sbjct: 75 ETAQAAKEKTGGAMQATKEKASEMGESAKETAVAGKEKTGGLMSSAAEQVKGMAQGATEA 134
Query: 58 VKSAIG 63
VK+ G
Sbjct: 135 VKNTFG 140
Score = 33.1 bits (74), Expect = 0.14
Identities = 17/62 (27%), Positives = 29/62 (46%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
M Q+ S++AG+ G AQEK M ++ AQ+A+ E + + + D S
Sbjct: 1 MASHQDQSYKAGEPKGHAQEKTGQMADTMKDKAQAAKDKASEMAGSARDRTVESKDQTGS 60
Query: 61 AI 62
+
Sbjct: 61 YV 62
>LEA3_MAIZE (Q42376) Late embryogenesis abundant protein, group 3
(LEA)
Length = 221
Score = 35.8 bits (81), Expect = 0.021
Identities = 19/61 (31%), Positives = 37/61 (60%), Gaps = 1/61 (1%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARN-AAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
+ ++ H+AG+ T A++KA A+ A ++ + + Q+AG+ +A Q AADA+++
Sbjct: 83 ETTEATKHKAGETTEAAKQKAGETTEAAKQKAGETTETTKQKAGETTEAARQKAADAMEA 142
Query: 61 A 61
A
Sbjct: 143 A 143
Score = 34.7 bits (78), Expect = 0.047
Identities = 16/52 (30%), Positives = 26/52 (49%)
Query: 8 SHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S+QAG+ + +EK +G ++ AQ A+ +A K Q A +A K
Sbjct: 9 SYQAGETKARTEEKTGQAVGATKDTAQHAKDRAADAAGHAAGKGQDAKEATK 60
Score = 34.3 bits (77), Expect = 0.062
Identities = 22/77 (28%), Positives = 37/77 (47%), Gaps = 15/77 (19%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSC---------------QEAGQQ 46
+ ++ +AG+ T A++KA++ M A+ A A Q Q+A +Q
Sbjct: 116 ETTETTKQKAGETTEAARQKAADAMEAAKQKAAEAGQYAKDTAVSGKDKSGGVIQQATEQ 175
Query: 47 MKAKAQGAADAVKSAIG 63
+K+ A G DAV S +G
Sbjct: 176 VKSAAAGRKDAVMSTLG 192
Score = 28.9 bits (63), Expect = 2.6
Identities = 23/80 (28%), Positives = 35/80 (43%), Gaps = 15/80 (18%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNAR----NAAQSAQQSCQEAGQQMKAKA------ 51
+ ++ +AG+ T ++KA AR +A ++A+Q EAGQ K A
Sbjct: 105 ETTEAAKQKAGETTETTKQKAGETTEAARQKAADAMEAAKQKAAEAGQYAKDTAVSGKDK 164
Query: 52 -----QGAADAVKSAIGANK 66
Q A + VKSA K
Sbjct: 165 SGGVIQQATEQVKSAAAGRK 184
>LEA3_WHEAT (Q03968) Late embryogenesis abundant protein, group 3
(LEA) (PMA2005)
Length = 224
Score = 34.7 bits (78), Expect = 0.047
Identities = 20/63 (31%), Positives = 37/63 (57%), Gaps = 2/63 (3%)
Query: 1 MDQSQN-LSHQAGQATGQAQEKASNMMGNARN-AAQSAQQSCQEAGQQMKAKAQGAADAV 58
M +QN S+ AG+ + +EK +MG ++ A Q+ + + Q+AG+ +A Q AA+
Sbjct: 1 MASNQNQASYHAGETKARNEEKTGQVMGATKDKAGQTTEATKQKAGETTEATKQKAAETT 60
Query: 59 KSA 61
++A
Sbjct: 61 EAA 63
Score = 28.1 bits (61), Expect = 4.4
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 4/58 (6%)
Query: 10 QAGQATGQAQEKASNMMGNARNAAQSAQQSC----QEAGQQMKAKAQGAADAVKSAIG 63
+A +AT A++KAS + +A + + Q+AG+ + GA DAV + +G
Sbjct: 139 KAAEATEAAKQKASETAQYTKESAVTGKDKTGSVLQQAGETVVNAVVGAKDAVANTLG 196
>LEA1_HORVU (P14928) ABA-inducible protein PHV A1
Length = 213
Score = 33.1 bits (74), Expect = 0.14
Identities = 19/63 (30%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query: 1 MDQSQNL-SHQAGQATGQAQEKASNMMG-NARNAAQSAQQSCQEAGQQMKAKAQGAADAV 58
M +QN S+ AG+ + +EK MMG + A Q+ + + Q+AG+ +A Q +
Sbjct: 1 MASNQNQGSYHAGETKARTEEKTGQMMGATKQKAGQTTEATKQKAGETAEATKQKTGETA 60
Query: 59 KSA 61
++A
Sbjct: 61 EAA 63
Score = 28.9 bits (63), Expect = 2.6
Identities = 17/66 (25%), Positives = 34/66 (50%), Gaps = 4/66 (6%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQS----CQEAGQQMKAKAQGAADA 57
+ ++ +A +AT A++KAS+ + +A + + Q+AG+ + GA DA
Sbjct: 120 ETTEAAKQKAAEATEAAKQKASDTAQYTKESAVAGKDKTGSVLQQAGETVVNAVVGAKDA 179
Query: 58 VKSAIG 63
V + +G
Sbjct: 180 VANTLG 185
>YK98_RHILO (Q98J52) Hypothetical UPF0337 protein mlr2098
Length = 71
Score = 31.6 bits (70), Expect = 0.40
Identities = 21/66 (31%), Positives = 31/66 (46%), Gaps = 4/66 (6%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGN----ARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S S A +A G+A+E +GN A AAQ A+ Q+A K+ + A +
Sbjct: 5 SDKASGLANEAVGKAKEGVGKAVGNDRLRAEGAAQEAKGKVQKAVGDAKSAVKDATNKTA 64
Query: 60 SAIGAN 65
+AI N
Sbjct: 65 AAINKN 70
>MYHB_RABIT (P35748) Myosin heavy chain, smooth muscle isoform (SMMHC)
Length = 1972
Score = 31.6 bits (70), Expect = 0.40
Identities = 21/57 (36%), Positives = 33/57 (57%), Gaps = 4/57 (7%)
Query: 7 LSHQAGQATGQAQEKASNMMGNARNAAQ---SAQQSCQEAGQQMKAKAQGAADAVKS 60
+S + +AT QA E+ SN + R+ AQ SA+Q + +++K+K Q AVKS
Sbjct: 1755 MSDRVRKATQQA-EQLSNELATERSTAQKNESARQQLERQNKELKSKLQEMEGAVKS 1810
>TOLA_ECOLI (P19934) TolA protein
Length = 421
Score = 30.8 bits (68), Expect = 0.68
Identities = 19/66 (28%), Positives = 36/66 (53%), Gaps = 2/66 (3%)
Query: 3 QSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQ--MKAKAQGAADAVKS 60
Q++ + QA QA+E A+ +A+ A++ ++ +EA ++ AK + A+A K+
Sbjct: 119 QAEEAAKQAELKQKQAEEAAAKAAADAKAKAEADAKAAEEAAKKAAADAKKKAEAEAAKA 178
Query: 61 AIGANK 66
A A K
Sbjct: 179 AAEAQK 184
>GALY_YEAST (P19659) Transcription regulatory protein GAL11
Length = 1081
Score = 30.4 bits (67), Expect = 0.89
Identities = 22/58 (37%), Positives = 27/58 (45%), Gaps = 1/58 (1%)
Query: 5 QNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQ-EAGQQMKAKAQGAADAVKSA 61
Q Q QA QAQ +A AAQ+AQ Q +A Q +A+AQ A A A
Sbjct: 416 QMQQQQQAQAQAQAQAQAQAQAQAQAQAAQAAQAQAQAQAQAQAQAQAQAQAQAQAQA 473
Score = 29.3 bits (64), Expect = 2.0
Identities = 24/64 (37%), Positives = 30/64 (46%), Gaps = 8/64 (12%)
Query: 1 MDQSQNLSHQA---GQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADA 57
M Q Q QA QA QAQ +A A AAQ+ Q+ +A Q +A+AQ A A
Sbjct: 417 MQQQQQAQAQAQAQAQAQAQAQAQA-----QAAQAAQAQAQAQAQAQAQAQAQAQAQAQA 471
Query: 58 VKSA 61
A
Sbjct: 472 QAQA 475
Score = 27.3 bits (59), Expect = 7.5
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 3/50 (6%)
Query: 3 QSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQ 52
Q+Q + QA QA QAQ +A A+ AQ+ Q+ +A Q +A AQ
Sbjct: 438 QAQAQAAQAAQAQAQAQAQA---QAQAQAQAQAQAQAQAQAQAQAQAHAQ 484
>AGL8_LYCES (Q40170) Agamous-like MADS box protein AGL8 homolog
(TM4)
Length = 227
Score = 30.4 bits (67), Expect = 0.89
Identities = 15/65 (23%), Positives = 28/65 (43%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
M + QNL HQ A + + + +M + + Q ++ QE Q+ K + + +
Sbjct: 122 MKELQNLEHQLDSALKHIRSRKNQLMHESISVLQKKDRALQEQNNQLSKKVKEREKSAQQ 181
Query: 61 AIGAN 65
G N
Sbjct: 182 ISGIN 186
>Y9D1_RHIME (Q92XV9) Hypothetical UPF0337 protein RA1131
Length = 63
Score = 29.3 bits (64), Expect = 2.0
Identities = 16/46 (34%), Positives = 24/46 (51%)
Query: 11 AGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAAD 56
AG+A A + N A+ AAQ A+ Q+A ++K +GA D
Sbjct: 16 AGKAKKAAGDATDNNSLRAKGAAQEAKGGAQQAKGKLKDAVKGAVD 61
>DRPF_CRAPL (P23283) Desiccation-related protein PCC3-06
Length = 201
Score = 28.9 bits (63), Expect = 2.6
Identities = 18/63 (28%), Positives = 25/63 (39%), Gaps = 11/63 (17%)
Query: 8 SHQAGQATGQAQEKASNM-----------MGNARNAAQSAQQSCQEAGQQMKAKAQGAAD 56
S +AG+ + QE A N+ M RNA + GQ +K GA +
Sbjct: 104 SGKAGELKDKTQEGAENVREKAMDAGNDAMEKTRNAGERVADGVSNVGQNVKENVMGAGE 163
Query: 57 AVK 59
VK
Sbjct: 164 KVK 166
Score = 28.1 bits (61), Expect = 4.4
Identities = 13/58 (22%), Positives = 26/58 (44%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
+ + +L H+ +AT A KA+ + A+ + ++K K Q A+ V+
Sbjct: 65 ETTDSLKHKTSEATDSASHKANGAARETNDKAKETYNAASGKAGELKDKTQEGAENVR 122
Score = 27.3 bits (59), Expect = 7.5
Identities = 16/66 (24%), Positives = 29/66 (43%), Gaps = 1/66 (1%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAA-DAVKS 60
+ + + SH+A A + +KA A A + QE + ++ KA A DA++
Sbjct: 76 EATDSASHKANGAARETNDKAKETYNAASGKAGELKDKTQEGAENVREKAMDAGNDAMEK 135
Query: 61 AIGANK 66
A +
Sbjct: 136 TRNAGE 141
>AGL8_SOLCO (O22328) Agamous-like MADS box protein AGL8 homolog
Length = 250
Score = 28.9 bits (63), Expect = 2.6
Identities = 13/52 (25%), Positives = 24/52 (46%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQ 52
M + QNL HQ A + + + +M + + Q ++ QE Q+ K +
Sbjct: 122 MKELQNLEHQLASALKHIRSRKNQLMHESISVLQKQDRALQEQNNQLSKKVK 173
>TPM_BRABE (Q9NDS0) Tropomyosin
Length = 284
Score = 28.5 bits (62), Expect = 3.4
Identities = 20/69 (28%), Positives = 31/69 (43%), Gaps = 10/69 (14%)
Query: 8 SHQAGQATGQAQEK----------ASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADA 57
+ QA QA AQEK + + + AQ+S +EA +Q++A + AADA
Sbjct: 22 AEQAEQAMKDAQEKNVKLEDEINDLNKKIRMVEDELDKAQESLKEATEQLEAATKKAADA 81
Query: 58 VKSAIGANK 66
N+
Sbjct: 82 EAEVASLNR 90
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.300 0.108 0.271
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,951,161
Number of Sequences: 164201
Number of extensions: 106889
Number of successful extensions: 527
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 448
Number of HSP's gapped (non-prelim): 84
length of query: 66
length of database: 59,974,054
effective HSP length: 42
effective length of query: 24
effective length of database: 53,077,612
effective search space: 1273862688
effective search space used: 1273862688
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0215b.3


