

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0212.5
(129 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
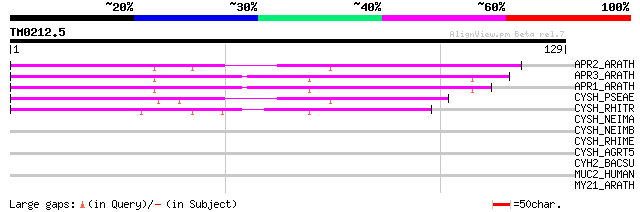
APR2_ARATH (P92981) 5'-adenylylsulfate reductase 2, chloroplast ... 93 9e-20
APR3_ARATH (P92980) 5'-adenylylsulfate reductase 3, chloroplast ... 91 3e-19
APR1_ARATH (P92979) 5'-adenylylsulfate reductase 1, chloroplast ... 90 8e-19
CYSH_PSEAE (O05927) Phosphoadenosine phosphosulfate reductase (E... 63 1e-10
CYSH_RHITR (O33579) Phosphoadenosine phosphosulfate reductase (E... 41 4e-04
CYSH_NEIMA (Q9JUD5) Phosphoadenosine phosphosulfate reductase (E... 38 0.004
CYSH_NEIMB (Q9JRT1) Phosphoadenosine phosphosulfate reductase (E... 36 0.017
CYSH_RHIME (P56891) Phosphoadenosine phosphosulfate reductase (E... 35 0.029
CYSH_AGRT5 (Q8UH67) Phosphoadenosine phosphosulfate reductase (E... 35 0.038
CYH2_BACSU (O06737) Probable phosphoadenosine phosphosulfate red... 34 0.065
MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2) 27 6.1
MY21_ARATH (Q9LK95) Transcription factor MYB21 (Myb-related prot... 27 7.9
>APR2_ARATH (P92981) 5'-adenylylsulfate reductase 2, chloroplast
precursor (EC 1.8.4.9) (Adenosine 5'-phosphosulfate
5'-adenylylsulfate sulfotransferase 2) (APS
sulfotransferase 2) (Thioredoxin independent APS
reductase 2) (3'-phosphoadenosine-5'-phosphos
Length = 454
Score = 93.2 bits (230), Expect = 9e-20
Identities = 62/157 (39%), Positives = 77/157 (48%), Gaps = 50/157 (31%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHERE 82
+A + A ++ +R P+ AL VL QHERE
Sbjct: 254 LANVEGADVWNF------------LRTMDVPVNALHAQGYVSIGCEPCTRPVLPGQHERE 301
Query: 83 GNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQV 119
G WWEDAKAK+CGLHKGN+K++ GA + A ++
Sbjct: 302 GRWWWEDAKAKECGLHKGNIKEEDGAADSKPAAVQEI 338
>APR3_ARATH (P92980) 5'-adenylylsulfate reductase 3, chloroplast
precursor (EC 1.8.4.9) (Adenosine 5'-phosphosulfate
5'-adenylylsulfate sulfotransferase 3) (APS
sulfotransferase 3) (Thioredoxin independent APS
reductase 3) (3'-phosphoadenosine-5'-phosphos
Length = 458
Score = 91.3 bits (225), Expect = 3e-19
Identities = 62/145 (42%), Positives = 75/145 (50%), Gaps = 30/145 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR+RKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 254 VANVEGNDVWNFLRT-MDVPVNTLHAAGYVSIGCEPCTRAVLPGQHEREGRWWWEDAKAK 312
Query: 94 DCGLHKGNVKQDV--GAQLNGNGAA 116
+CGLHKGN+K++ A N NG A
Sbjct: 313 ECGLHKGNIKENTNGNATANVNGTA 337
>APR1_ARATH (P92979) 5'-adenylylsulfate reductase 1, chloroplast
precursor (EC 1.8.4.9) (Adenosine 5'-phosphosulfate
5'-adenylylsulfate sulfotransferase 1) (APS
sulfotransferase 1) (Thioredoxin independent APS
reductase 1) (3'-phosphoadenosine-5'-phosphos
Length = 465
Score = 90.1 bits (222), Expect = 8e-19
Identities = 62/140 (44%), Positives = 75/140 (53%), Gaps = 29/140 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 202 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 261
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 262 VANVEGNDVWNFLRT-MDVPVNTLHAAGYISIGCEPCTKAVLPGQHEREGRWWWEDAKAK 320
Query: 94 DCGLHKGNVKQDV-GAQLNG 112
+CGLHKGNVK++ A++NG
Sbjct: 321 ECGLHKGNVKENSDDAKVNG 340
>CYSH_PSEAE (O05927) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 267
Score = 62.8 bits (151), Expect = 1e-10
Identities = 45/137 (32%), Positives = 60/137 (42%), Gaps = 47/137 (34%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGTN------------------LVKWN-LVA 41
CC +RK+ L+R+L GV GQ +DQSPGT L K+N L +
Sbjct: 139 CCGIRKIEPLKRKLAGVRAWATGQRRDQSPGTRSQVAVLEIDGAFSTPEKPLYKFNPLSS 198
Query: 42 ISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHEREGNR 85
++ ++G IR P +L VL QHEREG
Sbjct: 199 MTSEEVWGY------------IRMLELPYNSLHERGYISIGCEPCTRPVLPNQHEREGRW 246
Query: 86 WWEDAKAKDCGLHKGNV 102
WWE+A K+CGLH GN+
Sbjct: 247 WWEEATHKECGLHAGNL 263
>CYSH_RHITR (O33579) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase) (Fragment)
Length = 180
Score = 41.2 bits (95), Expect = 4e-04
Identities = 34/120 (28%), Positives = 53/120 (43%), Gaps = 27/120 (22%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQS------------PGTNLVKWNLVA-----IS 43
CC VRK++ L R L G T + G + QS P NL+K N +A +
Sbjct: 57 CCGVRKLKPLARALSGATIWVTGLRRGQSANRADTPFAEYDPERNLIKVNPLADWDIDVI 116
Query: 44 KAIIY--GISLGP*MCL*IH*IRKDTF---PLEALVLREQHEREGNRWWEDAKAKDCGLH 98
+A + G+ + P +H + P + + ER G WWE+ + ++CGLH
Sbjct: 117 RAYVADNGVPVNP-----LHQRGYPSIGCEPCTRAIKPGEPERAGRWWWENDEKRECGLH 171
>CYSH_NEIMA (Q9JUD5) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 244
Score = 37.7 bits (86), Expect = 0.004
Identities = 34/129 (26%), Positives = 51/129 (39%), Gaps = 43/129 (33%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSP------------GTNLVKWNLV-------- 40
CCR+RK L R + G + GQ ++QS G + K+N +
Sbjct: 129 CCRIRKTEPLDRAIAGADAWLTGQRREQSATRTELPFAEYDAGRGIDKYNPIFDWSEHDV 188
Query: 41 ---AISKAIIYGI-------SLGP*MCL*IH*IRKDTFPLEALVLREQHEREGNRWWEDA 90
++ + Y S+G C T P++A + R G WWED
Sbjct: 189 WAYILANNVPYNDLYRQGFPSIGCDPC---------TRPVKA----GEDIRAGRWWWEDK 235
Query: 91 KAKDCGLHK 99
+K+CGLHK
Sbjct: 236 NSKECGLHK 244
>CYSH_NEIMB (Q9JRT1) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 246
Score = 35.8 bits (81), Expect = 0.017
Identities = 33/129 (25%), Positives = 50/129 (38%), Gaps = 43/129 (33%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSP------------GTNLVKWNLV-------- 40
CCR+RK L R + G + GQ ++QS G + K+N +
Sbjct: 131 CCRIRKTEPLNRAIAGADAWLTGQRREQSATRTELPFAEYDAGRGIGKYNPIFDWSEHDV 190
Query: 41 ---AISKAIIYGI-------SLGP*MCL*IH*IRKDTFPLEALVLREQHEREGNRWWEDA 90
++ + Y S+G C T P++A + R G WWE
Sbjct: 191 WAYILANNVPYNDLYRQGFPSIGCDPC---------TRPVKA----GEDIRAGRWWWEGR 237
Query: 91 KAKDCGLHK 99
+K+CGLHK
Sbjct: 238 NSKECGLHK 246
>CYSH_RHIME (P56891) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 265
Score = 35.0 bits (79), Expect = 0.029
Identities = 31/120 (25%), Positives = 46/120 (37%), Gaps = 27/120 (22%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPG------------------TNLVKWNLVAI 42
CC VRK++ L R L G + I G + QS L W + I
Sbjct: 135 CCGVRKLKPLARALDGASYWITGLRRGQSGNRATTPFAEADVERGLIKINPLADWGIETI 194
Query: 43 SKAIIY-GISLGP*MCL*IH*IRKDTF---PLEALVLREQHEREGNRWWEDAKAKDCGLH 98
+ GI + P +H + P + + ER G WWE+ + ++CGLH
Sbjct: 195 QAHVAAEGIPVNP-----LHSRGYPSIGCEPCTRAIKPGEPERAGRWWWENDEKRECGLH 249
>CYSH_AGRT5 (Q8UH67) Phosphoadenosine phosphosulfate reductase (EC
1.8.4.8) (PAPS reductase, thioredoxin dependent) (PAdoPS
reductase) (3'-phosphoadenylylsulfate reductase) (PAPS
sulfotransferase)
Length = 260
Score = 34.7 bits (78), Expect = 0.038
Identities = 34/126 (26%), Positives = 51/126 (39%), Gaps = 27/126 (21%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT------------NLVKWNLVA---ISKA 45
CC VRK+ L + L+G I G + QS NL+K N +A I +
Sbjct: 130 CCHVRKLIPLGKALEGAAFWITGLRRGQSGNRAATPFAEFDAERNLIKINALADWDIEQI 189
Query: 46 IIY----GISLGP*MCL*IH*IRKDTF---PLEALVLREQHEREGNRWWEDAKAKDCGLH 98
Y I + P +H + P + + ER G WWE+ + ++CGLH
Sbjct: 190 RAYVAEENIPVNP-----LHQRGYPSIGCEPCTRAIKPGEPERAGRWWWENDEKRECGLH 244
Query: 99 KGNVKQ 104
+Q
Sbjct: 245 VAGAEQ 250
>CYH2_BACSU (O06737) Probable phosphoadenosine phosphosulfate
reductase (EC 1.8.4.8) (PAPS reductase, thioredoxin
dependent) (PAdoPS reductase) (3'-phosphoadenylylsulfate
reductase) (PAPS sulfotransferase)
Length = 236
Score = 33.9 bits (76), Expect = 0.065
Identities = 14/30 (46%), Positives = 21/30 (69%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSP 30
CC++RK+ L++ L G+T I G +DQSP
Sbjct: 122 CCQLRKIEPLKKHLSGMTAWISGLRRDQSP 151
>MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2)
Length = 5179
Score = 27.3 bits (59), Expect = 6.1
Identities = 17/57 (29%), Positives = 23/57 (39%), Gaps = 15/57 (26%)
Query: 70 LEALVLREQHEREGNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQVHCHCRIH 126
LE L E+H +G C +G V D+G +G CHCR+H
Sbjct: 315 LEVSSLCEEHRMDG-----------CFCPEGTVYDDIG----DSGCVPVSQCHCRLH 356
>MY21_ARATH (Q9LK95) Transcription factor MYB21 (Myb-related protein
21) (AtMYB21) (Myb homolog 3) (AtMyb3)
Length = 226
Score = 26.9 bits (58), Expect = 7.9
Identities = 12/28 (42%), Positives = 17/28 (59%)
Query: 64 RKDTFPLEALVLREQHEREGNRWWEDAK 91
R + P E L++ E H + GNRW + AK
Sbjct: 75 RGNITPEEQLIIMELHAKWGNRWSKIAK 102
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.344 0.153 0.534
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,456,834
Number of Sequences: 164201
Number of extensions: 469427
Number of successful extensions: 1572
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 1550
Number of HSP's gapped (non-prelim): 19
length of query: 129
length of database: 59,974,054
effective HSP length: 105
effective length of query: 24
effective length of database: 42,732,949
effective search space: 1025590776
effective search space used: 1025590776
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0212.5


