

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0211.5
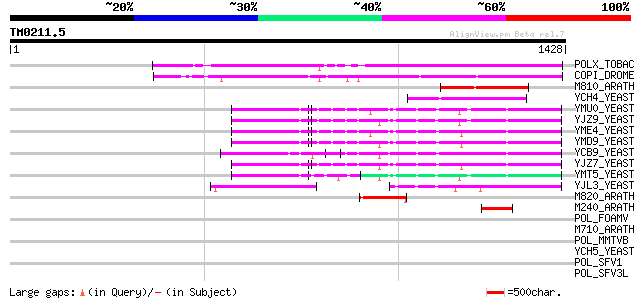
(1428 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 508 e-143
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 469 e-131
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 198 1e-49
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 164 1e-39
YMU0_YEAST (Q04670) Transposon Ty1 protein B 137 3e-31
YJZ9_YEAST (P47100) Transposon Ty1 protein B 136 5e-31
YME4_YEAST (Q04711) Transposon Ty1 protein B 134 1e-30
YMD9_YEAST (Q03434) Transposon Ty1 protein B 133 4e-30
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 132 5e-30
YJZ7_YEAST (P47098) Transposon Ty1 protein B 130 2e-29
YMT5_YEAST (Q04214) Transposon Ty1 protein B 129 4e-29
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 126 4e-28
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 112 7e-24
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 97 3e-19
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 42 0.012
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 42 0.012
POL_MMTVB (P03365) Pol polyprotein [Contains: Reverse transcript... 41 0.027
YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein 40 0.046
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 40 0.060
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 39 0.078
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 508 bits (1309), Expect = e-143
Identities = 363/1095 (33%), Positives = 535/1095 (48%), Gaps = 98/1095 (8%)
Query: 367 DTTWILDTGATDHICNTLSYFSSYKHVEPIPVSLPNGIVETTTIKGTIQITPS----FIL 422
++ W++DT A+ H F Y + V + N G I I + +L
Sbjct: 291 ESEWVVDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVL 350
Query: 423 ANVLFLPNFEFNLISVHKLVKCLRYRLIFEDDLCLIQDSNACKMIGTVRAVKGLYIFNKS 482
+V +P+ NLIS L + Y F + + + G R LY N
Sbjct: 351 KDVRHVPDLRMNLISGIALDRD-GYESYFANQKWRLTKGSLVIAKGVARGT--LYRTNAE 407
Query: 483 SISSLASCNSISTSVNPSVHSSSICTFQSNVHNLWHYRLGHPSLVKGQSI---NELFPYV 539
+ SV+ LWH R+GH S KG I L Y
Sbjct: 408 ICQGELNAAQDEISVD-----------------LWHKRMGHMS-EKGLQILAKKSLISYA 449
Query: 540 QCSKAHVCDVCPVAKQKRMSFPLSVTQSTAIFQLIHVDIWGPVSIVSLHGFSYFLTIVDD 599
+ + CD C KQ R+SF S + I L++ D+ GP+ I S+ G YF+T +DD
Sbjct: 450 KGTTVKPCDYCLFGKQHRVSFQTSSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDD 509
Query: 600 YSRFTWIYLLKS*AEVKNLVQEFCALVANQFETAVKTIRSDNGKEFSLPQFY---ATKGI 656
SR W+Y+LK+ +V + Q+F ALV + +K +RSDNG E++ +F ++ GI
Sbjct: 510 ASRKLWVYILKTKDQVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEYCSSHGI 569
Query: 657 VHQTSCVETPQQNSIVERKHQHILNVARALLFQAHLPKIFWAHAIVHAVFLINRLPSPVL 716
H+ + TPQ N + ER ++ I+ R++L A LPK FW A+ A +LINR PS L
Sbjct: 570 RHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACYLINRSPSVPL 629
Query: 717 DGKCPFQILHKVLPDLTNLKVFGSLCFASTLVSHRTKFDPRAKRCVFLGFKPGTKGYIVY 776
+ P ++ ++LKVFG FA RTK D ++ C+F+G+ GY ++
Sbjct: 630 AFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLW 689
Query: 777 DLKSNDIAISRNVVFHEN---------------MFP-YPTQPSDLNQHQSCPLPQASFLS 820
D + SR+VVF E+ + P + T PS N S A +
Sbjct: 690 DPVKKKVIRSRDVVFRESEVRTAADMSEKVKNGIIPNFVTIPSTSNNPTS-----AESTT 744
Query: 821 DEPFEYATQSPSEALTQPNTSEPSSDPVLDNNHRTSTRTRKQPSYLQDYHCSLIASTAVS 880
DE E Q P E + Q E + V + H T + QP S
Sbjct: 745 DEVSEQGEQ-PGEVIEQ---GEQLDEGVEEVEHPTQGEEQHQP-------------LRRS 787
Query: 881 SSSSSKGTSYPLSKVISYCNLAPAYHTFVMNITAVVEPKRYSEAVKH---DSWRKAMDQE 937
+ YP ++ + I+ EP+ E + H + KAM +E
Sbjct: 788 ERPRVESRRYPSTEYVL--------------ISDDREPESLKEVLSHPEKNQLMKAMQEE 833
Query: 938 IEALERNHTWILVDKPHDKTPIGCKWVYRIKYKQDGTLDRYKARLVVKGYTQLEGIDFID 997
+E+L++N T+ LV+ P K P+ CKWV+++K D L RYKARLVVKG+ Q +GIDF +
Sbjct: 834 MESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFDE 893
Query: 998 TFSPVAKMTTLRVLLALASSYNWFLHQLDVDNAFLHAQLDEEIYMSLPQGLHTE-KPNQV 1056
FSPV KMT++R +L+LA+S + + QLDV AFLH L+EEIYM P+G K + V
Sbjct: 894 IFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHMV 953
Query: 1057 CLLQKSLYGLKQASRQWYTTLCKALHTLGFSPSSADHTLYIKKGTTGSFTALLLYVDDVL 1116
C L KSLYGLKQA RQWY + + + + +D +Y K+ + +F LLLYVDD+L
Sbjct: 954 CKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDML 1013
Query: 1117 LTGNDLHEIQLVKESLHAQFRIKDMGEAKFFLGLEIARSKAG--IVLNQRKYALELLSDS 1174
+ G D I +K L F +KD+G A+ LG++I R + + L+Q KY +L
Sbjct: 1014 IVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLERF 1073
Query: 1175 GLLGGKPTTTPMDS----SQKFGLSTDTPLSDISS--YRRLIGKLLY-LTTTRPDIAYVV 1227
+ KP +TP+ S+K +T +++ Y +G L+Y + TRPDIA+ V
Sbjct: 1074 NMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTRPDIAHAV 1133
Query: 1228 NQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLFYPADSSTTLTAFSDSDWAGCLDTRKS 1287
+S+FL P H A +LRY++G G L + S L ++D+D AG +D RKS
Sbjct: 1134 GVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCF-GGSDPILKGYTDADMAGDIDNRKS 1192
Query: 1288 ITGYCMFLGSSLISWRSKKQTTTSRSSCEAEYRAMAATVCEVQWLSYLLHDLQAPPSAPV 1347
TGY ISW+SK Q + S+ EAEY A T E+ WL L +L V
Sbjct: 1193 STGYLFTFSGGAISWQSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGLHQKEYV 1252
Query: 1348 SMYCDNQSAMHIAHNPSYHERTKHIEVDCHIVREKVQQGLVHLLPIASSHQLADIFTKPL 1407
+YCD+QSA+ ++ N YH RTKHI+V H +RE V + +L I+++ AD+ TK +
Sbjct: 1253 -VYCDSQSAIDLSKNSMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPADMLTKVV 1311
Query: 1408 TPAPFRHIFSKLGMY 1422
F +GM+
Sbjct: 1312 PRNKFELCKELVGMH 1326
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 469 bits (1206), Expect = e-131
Identities = 350/1144 (30%), Positives = 550/1144 (47%), Gaps = 124/1144 (10%)
Query: 370 WILDTGATDHICNTLS-YFSSYKHVEPIPVSLPN-GIVETTTIKGTIQITPS--FILANV 425
++LD+GA+DH+ N S Y S + V P+ +++ G T +G +++ L +V
Sbjct: 289 FVLDSGASDHLINDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGIVRLRNDHEITLEDV 348
Query: 426 LFLPNFEFNLISVHKLVKCLRYRLIFEDDLCLIQDSNACKMIGTVRAVKGLYIF-NKSSI 484
LF NL+SV +L E + + D + G + GL + N +
Sbjct: 349 LFCKEAAGNLMSVKRLQ---------EAGMSIEFDKS-----GVTISKNGLMVVKNSGML 394
Query: 485 SSLASCNSISTSVNPSVHSSSICTFQSNVHNLWHYRLGHPSLVKGQSINELFPYVQCS-- 542
+++ N + S+N N LWH R GH S K I + S
Sbjct: 395 NNVPVINFQAYSINAK---------HKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLL 445
Query: 543 -----KAHVCDVCPVAKQKRMSFPLSVTQSTAIFQ---LIHVDIWGPVSIVSLHGFSYFL 594
+C+ C KQ R+ F + T I + ++H D+ GP++ V+L +YF+
Sbjct: 446 NNLELSCEICEPCLNGKQARLPFK-QLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFV 504
Query: 595 TIVDDYSRFTWIYLLKS*AEVKNLVQEFCALVANQFETAVKTIRSDNGKEF---SLPQFY 651
VD ++ + YL+K ++V ++ Q+F A F V + DNG+E+ + QF
Sbjct: 505 IFVDQFTHYCVTYLIKYKSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFC 564
Query: 652 ATKGIVHQTSCVETPQQNSIVERKHQHILNVARALLFQAHLPKIFWAHAIVHAVFLINRL 711
KGI + + TPQ N + ER + I AR ++ A L K FW A++ A +LINR+
Sbjct: 565 VKKGISYHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRI 624
Query: 712 PSPVL--DGKCPFQILHKVLPDLTNLKVFGSLCFASTLVSHRTKFDPRAKRCVFLGFKPG 769
PS L K P+++ H P L +L+VFG+ + + + + KFD ++ + +F+G++P
Sbjct: 625 PSRALVDSSKTPYEMWHNKKPYLKHLRVFGATVYVH-IKNKQGKFDDKSFKSIFVGYEPN 683
Query: 770 TKGYIVYDLKSNDIAISRNVVFHE-NM-----------------------FPYPT----Q 801
G+ ++D + ++R+VV E NM FP + Q
Sbjct: 684 --GFKLWDAVNEKFIVARDVVVDETNMVNSRAVKFETVFLKDSKESENKNFPNDSRKIIQ 741
Query: 802 PSDLNQHQSCPLPQASFLSDEPFEYATQSPSEALTQPNTSEPSSDPVLDNNHRTSTRTRK 861
N+ + C Q FL D P+++ T P+ DN
Sbjct: 742 TEFPNESKECDNIQ--FLKDSKESENKNFPNDSRKIIQTEFPNESKECDNIQFLKDSKES 799
Query: 862 QPSYL-------QDYHCSLIASTAVSSSSSSKGTSYPLSKV------------------- 895
+L +D H + + + S T+ L ++
Sbjct: 800 NKYFLNESKKRKRDDHLNESKGSGNPNESRESETAEHLKEIGIDNPTKNDGIEIINRRSE 859
Query: 896 -------ISYCNLAPAYHTFVMNITAVVE--PKRYSEAVKHD---SWRKAMDQEIEALER 943
ISY + + V+N + P + E D SW +A++ E+ A +
Sbjct: 860 RLKTKPQISYNEEDNSLNKVVLNAHTIFNDVPNSFDEIQYRDDKSSWEEAINTELNAHKI 919
Query: 944 NHTWILVDKPHDKTPIGCKWVYRIKYKQDGTLDRYKARLVVKGYTQLEGIDFIDTFSPVA 1003
N+TW + +P +K + +WV+ +KY + G RYKARLV +G+TQ ID+ +TF+PVA
Sbjct: 920 NNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQKYQIDYEETFAPVA 979
Query: 1004 KMTTLRVLLALASSYNWFLHQLDVDNAFLHAQLDEEIYMSLPQGLHTEKPNQVCLLQKSL 1063
++++ R +L+L YN +HQ+DV AFL+ L EEIYM LPQG+ N VC L K++
Sbjct: 980 RISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGISCNSDN-VCKLNKAI 1038
Query: 1064 YGLKQASRQWYTTLCKALHTLGFSPSSADHTLYI-KKGTTGSFTALLLYVDDVLLTGNDL 1122
YGLKQA+R W+ +AL F SS D +YI KG +LLYVDDV++ D+
Sbjct: 1039 YGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYVLLYVDDVVIATGDM 1098
Query: 1123 HEIQLVKESLHAQFRIKDMGEAKFFLGLEIARSKAGIVLNQRKYALELLSDSGLLGGKPT 1182
+ K L +FR+ D+ E K F+G+ I + I L+Q Y ++LS +
Sbjct: 1099 TRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFNMENCNAV 1158
Query: 1183 TTPMDSSQKFG-LSTDTPLSDISSYRRLIGKLLY-LTTTRPDIAYVVNQLSQFLSAPTNV 1240
+TP+ S + L++D + + R LIG L+Y + TRPD+ VN LS++ S +
Sbjct: 1159 STPLPSKINYELLNSDEDCN--TPCRSLIGCLMYIMLCTRPDLTTAVNILSRYSSKNNSE 1216
Query: 1241 HEAAAHRVLRYIKGNPGCGLFYPADSS--TTLTAFSDSDWAGCLDTRKSITGYCM-FLGS 1297
RVLRY+KG L + + + + + DSDWAG RKS TGY
Sbjct: 1217 LWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSEIDRKSTTGYLFKMFDF 1276
Query: 1298 SLISWRSKKQTTTSRSSCEAEYRAMAATVCEVQWLSYLLHDLQAPPSAPVSMYCDNQSAM 1357
+LI W +K+Q + + SS EAEY A+ V E WL +LL + P+ +Y DNQ +
Sbjct: 1277 NLICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFLLTSINIKLENPIKIYEDNQGCI 1336
Query: 1358 HIAHNPSYHERTKHIEVDCHIVREKVQQGLVHLLPIASSHQLADIFTKPLTPAPFRHIFS 1417
IA+NPS H+R KHI++ H RE+VQ ++ L I + +QLADIFTKPL A F +
Sbjct: 1337 SIANNPSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTENQLADIFTKPLPAARFVELRD 1396
Query: 1418 KLGM 1421
KLG+
Sbjct: 1397 KLGL 1400
Score = 35.4 bits (80), Expect = 1.1
Identities = 53/269 (19%), Positives = 101/269 (36%), Gaps = 50/269 (18%)
Query: 6 YHNWARAMKMSLLTKNKLGFVDGTIAEPPQDHPVLPFWQRCNMLVLSWLIKSISVEIAQS 65
Y W ++ L ++ L VDG + D W++ S +I+ +S
Sbjct: 16 YAIWKFRIRALLAEQDVLKVVDGLMPNEVDDS-----WKKAERCAKSTIIEYLSDSFLNF 70
Query: 66 ILWRDKATDVWNELRERFAQADLFRISELQEEIFSLKQG------------DNSVSKFYT 113
A + L + + L L++ + SLK D +S+
Sbjct: 71 ATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFHIFDELISELLA 130
Query: 114 SMKTLWDELDILNPLPVCTCNPRCACGAIKNIEDERNKNQVVRFLRGLNDQFSGVRSQLM 173
+ + +E+D ++ L + P C G I IE +N + F++
Sbjct: 131 AGAKI-EEMDKISHLLITL--PSCYDGIITAIETLSEENLTLAFVK-------------- 173
Query: 174 LLDNLPNVNRVFALIAQQERQFSFENVSGSRALIASRENSNDNRGSQSDHNRNSQSNYGG 233
NR L+ Q+ + + N + + + A N+N+ + NR ++
Sbjct: 174 --------NR---LLDQEIKIKNDHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPK--- 219
Query: 234 CQSSSGNNRYSSKKCSYCGKMGHTVEDCY 262
+ GN++Y K C +CG+ GH +DC+
Sbjct: 220 -KIFKGNSKYKVK-CHHCGREGHIKKDCF 246
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 198 bits (503), Expect = 1e-49
Identities = 102/226 (45%), Positives = 143/226 (63%), Gaps = 1/226 (0%)
Query: 1108 LLLYVDDVLLTGNDLHEIQLVKESLHAQFRIKDMGEAKFFLGLEIARSKAGIVLNQRKYA 1167
LLLYVDD+LLTG+ + ++ L + F +KD+G +FLG++I +G+ L+Q KYA
Sbjct: 3 LLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYA 62
Query: 1168 LELLSDSGLLGGKPTTTPMDSSQKFGLSTDTPLSDISSYRRLIGKLLYLTTTRPDIAYVV 1227
++L+++G+L KP +TP+ +ST D S +R ++G L YLT TRPDI+Y V
Sbjct: 63 EQILNNAGMLDCKPMSTPLPLKLNSSVST-AKYPDPSDFRSIVGALQYLTLTRPDISYAV 121
Query: 1228 NQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLFYPADSSTTLTAFSDSDWAGCLDTRKS 1287
N + Q + PT RVLRY+KG GL+ +S + AF DSDWAGC TR+S
Sbjct: 122 NIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRRS 181
Query: 1288 ITGYCMFLGSSLISWRSKKQTTTSRSSCEAEYRAMAATVCEVQWLS 1333
TG+C FLG ++ISW +K+Q T SRSS E EYRA+A T E+ W S
Sbjct: 182 TTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTWSS 227
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 164 bits (416), Expect = 1e-39
Identities = 108/309 (34%), Positives = 161/309 (51%), Gaps = 5/309 (1%)
Query: 1025 LDVDNAFLHAQLDEEIYMSLPQGLHTEK-PNQVCLLQKSLYGLKQASRQWYTTLCKALHT 1083
+DVD AFL++ +DE IY+ P G E+ P+ V L +YGLKQA W + L
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 1084 LGFSPSSADHTLYIKKGTTGSFTALLLYVDDVLLTGNDLHEIQLVKESLHAQFRIKDMGE 1143
+GF +H LY + + G + +YVDD+L+ VK+ L + +KD+G+
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIY-IGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGK 119
Query: 1144 AKFFLGLEIARSKAG-IVLNQRKYALELLSDSGLLGGKPTTTPMDSSQKFGLSTDTPLSD 1202
FLGL I +S G I L+ + Y + S+S + K T TP+ +S+ +T L D
Sbjct: 120 VDKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKD 179
Query: 1203 ISSYRRLIGKLLYLTTT-RPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLF 1261
I+ Y+ ++G+LL+ T RPDI+Y V+ LS+FL P +H +A RVLRY+ L
Sbjct: 180 ITPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLK 239
Query: 1262 YPADSSTTLTAFSDSDWAGCLDTRKSITGYCMFLGSSLISWRSKK-QTTTSRSSCEAEYR 1320
Y + S LT + D+ D S GY L + ++W SKK + S EAEY
Sbjct: 240 YRSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYI 299
Query: 1321 AMAATVCEV 1329
+ TV E+
Sbjct: 300 TASETVMEI 308
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 137 bits (344), Expect = 3e-31
Identities = 172/701 (24%), Positives = 294/701 (41%), Gaps = 85/701 (12%)
Query: 770 TKGYIVYDLKSNDIAISRNVVFHENMFPY--PTQPSDLNQHQS--CPLPQASFLSDEPFE 825
T+ I++ S D + N H N+ P PT S+ N +S LP + P E
Sbjct: 658 TEKRIIHRSPSIDASPPENNSSH-NIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTE 716
Query: 826 YATQSPSEALTQPNTSEPSSD--PVLDNNHRTSTRTRKQPSYLQDYHCSLIASTAVSSSS 883
+ P + L N+ + +S + D+N T+ ++K+ L+D + S ++
Sbjct: 717 FP--DPFKELPPINSRQTNSSLGGIGDSNAYTTINSKKRS--LEDNETEIKVSRDTWNTK 772
Query: 884 SSKGTSYPLSKVISYCNLAPAYHTFVMNITAVVEPKRYSEAVKH-------DSWRKAMDQ 936
+ + P SK +L A V +I + RY EA+ + + + +A +
Sbjct: 773 NMRSLEPPRSK--KRIHLIAAVKA-VKSIKPIRTTLRYDEAITYNKDIKEKEKYIQAYHK 829
Query: 937 EIEALERNHTWILVDKPHDKTPIGCKWVYRIKY----KQDGTLDRYKARLVVKGYTQLEG 992
E+ L + TW D+ +D+ I K V + K+DGT +KAR V +G
Sbjct: 830 EVNQLLKMKTWD-TDRYYDRKEIDPKRVINSMFIFNRKRDGT---HKARFVARG-----D 880
Query: 993 IDFIDTFSPVAKMTT-----LRVLLALASSYNWFLHQLDVDNAFLHAQLDEEIYMSLPQG 1047
I DT+ P + T L L+LA N+++ QLD+ +A+L+A + EE+Y+ P
Sbjct: 881 IQHPDTYDPGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP- 939
Query: 1048 LHTEKPNQVCLLQKSLYGLKQASRQWYTTLCKALHTLGFSPSSADHTLYIKKGTTGSFTA 1107
H +++ L+KSLYGLKQ+ WY T+ L + K S
Sbjct: 940 -HLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFK----NSQVT 994
Query: 1108 LLLYVDDVLLTGNDLHEIQLVKESLHAQF--RIKDMGEAK-----FFLGLEIARS----- 1155
+ L+VDD++L DL+ + + +L Q+ +I ++GE+ LGLEI
Sbjct: 995 ICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYM 1054
Query: 1156 -------------KAGIVLNQRKYALELLSDSGLLGGKPTTTPMDSSQKFGLSTDTPLSD 1202
K + LN + L GL Q+ L D
Sbjct: 1055 KLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGL---------YIDQQELELEEDDYKMK 1105
Query: 1203 ISSYRRLIGKLLYL-TTTRPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLF 1261
+ ++LIG Y+ R D+ Y +N L+Q + P+ + ++++I L
Sbjct: 1106 VHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLI 1165
Query: 1262 Y----PADSSTTLTAFSDSDWAGCLDTRKSITGYCMFLGSSLISWRSKKQTTTSRSSCEA 1317
+ P + L SD+ + G KS G L +I +S K + T S+ EA
Sbjct: 1166 WHKSKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEA 1224
Query: 1318 EYRAMAATVCEVQWLSYLLHDLQAPPSAPVSMYCDNQSAMHIAHNPSYHERTKHIEVDCH 1377
E A++ +V + LS+L+ +L P + + I N R +
Sbjct: 1225 EIHAISESVPLLNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAM 1284
Query: 1378 IVREKVQQGLVHLLPIASSHQLADIFTKPLTPAPFRHIFSK 1418
+R++V +H+ I + +AD+ TKPL F+ + +K
Sbjct: 1285 RLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
Score = 74.3 bits (181), Expect = 2e-12
Identities = 58/211 (27%), Positives = 95/211 (44%), Gaps = 7/211 (3%)
Query: 571 FQLIHVDIWGPVSIVSLHGFSYFLTIVDDYSRFTWIYLLKS*AE--VKNLVQEFCALVAN 628
FQ +H DI+GPV + SYF++ D+ ++F W+Y L E + ++ A + N
Sbjct: 238 FQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKN 297
Query: 629 QFETAVKTIRSDNGKEF---SLPQFYATKGIVHQTSCVETPQQNSIVERKHQHILNVARA 685
QF+ +V I+ D G E+ +L +F GI + + + + ER ++ +L+ R
Sbjct: 298 QFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRT 357
Query: 686 LLFQAHLPKIFWAHAIVHAVFLINRLPSPVLDGKCPFQILHKVLPDLTNLKVFGSLCFAS 745
L + LP W AI + + N L SP K Q D++ L FG +
Sbjct: 358 QLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVIVN 416
Query: 746 TLVSHRTKFDPRAKRCVFLGFKPGTKGYIVY 776
+ +K PR L + GYI+Y
Sbjct: 417 D-HNPNSKIHPRGIPGYALHPSRNSYGYIIY 446
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 136 bits (342), Expect = 5e-31
Identities = 172/701 (24%), Positives = 293/701 (41%), Gaps = 85/701 (12%)
Query: 770 TKGYIVYDLKSNDIAISRNVVFHENMFPY--PTQPSDLNQHQS--CPLPQASFLSDEPFE 825
T+ I++ S D + N H N+ P PT S+ N +S LP + P E
Sbjct: 1085 TEKRIIHRSPSIDASPPENNSSH-NIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTE 1143
Query: 826 YATQSPSEALTQPNTSEPSSD--PVLDNNHRTSTRTRKQPSYLQDYHCSLIASTAVSSSS 883
+ P + L N+ + +S + D+N T+ ++K+ L+D + S ++
Sbjct: 1144 FP--DPFKELPPINSRQTNSSLGGIGDSNAYTTINSKKRS--LEDNETEIKVSRDTWNTK 1199
Query: 884 SSKGTSYPLSKVISYCNLAPAYHTFVMNITAVVEPKRYSEAVKHDSWRKAMDQEIEALER 943
+ + P SK +L A V +I + RY EA+ ++ K ++ IEA +
Sbjct: 1200 NMRSLEPPRSK--KRIHLIAAVKA-VKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHK 1256
Query: 944 NHTWIL------VDKPHDKTPIGCKWVYRIKY----KQDGTLDRYKARLVVKGYTQLEGI 993
+L D+ +D+ I K V + K+DGT +KAR V +G I
Sbjct: 1257 EVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKKRDGT---HKARFVARG-----DI 1308
Query: 994 DFIDTFSPVAKMTT-----LRVLLALASSYNWFLHQLDVDNAFLHAQLDEEIYMSLPQGL 1048
DT+ + T L L+LA N+++ QLD+ +A+L+A + EE+Y+ P
Sbjct: 1309 QHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP-- 1366
Query: 1049 HTEKPNQVCLLQKSLYGLKQASRQWYTTLCKAL-HTLGFSPSSADHTLYIKKGTTGSFTA 1107
H +++ L+KSLYGLKQ+ WY T+ L G ++ S
Sbjct: 1367 HLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIQQCGMEEVRGWSCVF-----KNSQVT 1421
Query: 1108 LLLYVDDVLLTGNDLHEIQLVKESLHAQF--RIKDMGEAK-----FFLGLEIARS----- 1155
+ L+VDD++L +L+ + + E L Q+ +I ++GE+ LGLEI
Sbjct: 1422 ICLFVDDMVLFSKNLNSNKRIIEKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYM 1481
Query: 1156 -------------KAGIVLNQRKYALELLSDSGLLGGKPTTTPMDSSQKFGLSTDTPLSD 1202
K + LN + L GL Q+ L D
Sbjct: 1482 KLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGL---------YIDQQELELEEDDYKMK 1532
Query: 1203 ISSYRRLIGKLLYL-TTTRPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLF 1261
+ ++LIG Y+ R D+ Y +N L+Q + P+ + ++++I L
Sbjct: 1533 VHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLI 1592
Query: 1262 Y----PADSSTTLTAFSDSDWAGCLDTRKSITGYCMFLGSSLISWRSKKQTTTSRSSCEA 1317
+ P + L SD+ + G KS G L +I +S K + T S+ EA
Sbjct: 1593 WHKSKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEA 1651
Query: 1318 EYRAMAATVCEVQWLSYLLHDLQAPPSAPVSMYCDNQSAMHIAHNPSYHERTKHIEVDCH 1377
E A++ +V + LSYL+ +L P + + I N R +
Sbjct: 1652 EIHAISESVPLLNNLSYLIQELDKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAM 1711
Query: 1378 IVREKVQQGLVHLLPIASSHQLADIFTKPLTPAPFRHIFSK 1418
+R++V +H+ I + +AD+ TKPL F+ + +K
Sbjct: 1712 RLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1752
Score = 74.3 bits (181), Expect = 2e-12
Identities = 58/211 (27%), Positives = 95/211 (44%), Gaps = 7/211 (3%)
Query: 571 FQLIHVDIWGPVSIVSLHGFSYFLTIVDDYSRFTWIYLLKS*AE--VKNLVQEFCALVAN 628
FQ +H DI+GPV + SYF++ D+ ++F W+Y L E + ++ A + N
Sbjct: 665 FQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKN 724
Query: 629 QFETAVKTIRSDNGKEF---SLPQFYATKGIVHQTSCVETPQQNSIVERKHQHILNVARA 685
QF+ +V I+ D G E+ +L +F GI + + + + ER ++ +L+ R
Sbjct: 725 QFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRT 784
Query: 686 LLFQAHLPKIFWAHAIVHAVFLINRLPSPVLDGKCPFQILHKVLPDLTNLKVFGSLCFAS 745
L + LP W AI + + N L SP K Q D++ L FG +
Sbjct: 785 QLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVIVN 843
Query: 746 TLVSHRTKFDPRAKRCVFLGFKPGTKGYIVY 776
+ +K PR L + GYI+Y
Sbjct: 844 D-HNPNSKIHPRGIPGYALHPSRNSYGYIIY 873
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 134 bits (338), Expect = 1e-30
Identities = 170/692 (24%), Positives = 299/692 (42%), Gaps = 67/692 (9%)
Query: 770 TKGYIVYDLKSNDIAISRNVVFHENMFPY--PTQPSDLNQHQS--CPLPQASFLSDEPFE 825
T+ I++ S D + N H N+ P PT S+ N +S LP + P E
Sbjct: 658 TEKRIIHRSPSIDASPPENNSSH-NIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTE 716
Query: 826 YATQSPSEALTQPNTSEPSSD--PVLDNNHRTSTRTRKQPSYLQDYHCSLIASTAVSSSS 883
+ P + L N+ + +S + D+N T+ ++K+ L+D + S ++
Sbjct: 717 FP--DPFKELPPINSHQTNSSLGGIGDSNAYTTINSKKRS--LEDNETEIKVSRDTWNTK 772
Query: 884 SSKGTSYPLSKVISYCNLAPAYHTFVMNITAVVEPKRYSEAVKH-------DSWRKAMDQ 936
+ + P SK +L A V +I + RY EA+ + + + +A +
Sbjct: 773 NMRSLEPPRSK--KRIHLIAAVKA-VKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHK 829
Query: 937 EIEALERNHTWILVDKPHDKTPIGCKWVYRIKY----KQDGTLDRYKARLVVKGYTQLEG 992
E+ L + +TW DK +D+ I K V + K+DGT +KAR V +G
Sbjct: 830 EVNQLLKMNTWD-TDKYYDRKEIDPKRVINSMFIFNRKRDGT---HKARFVARG-----D 880
Query: 993 IDFIDTFSPVAKMTT-----LRVLLALASSYNWFLHQLDVDNAFLHAQLDEEIYMSLPQG 1047
I DT+ + T L L+LA N+++ QLD+ +A+L+A + EE+Y+ P
Sbjct: 881 IQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP- 939
Query: 1048 LHTEKPNQVCLLQKSLYGLKQASRQWYTTLCKALHTLGFSPSSADHTLYIKKGTTGSFTA 1107
H +++ L+KSLYGLKQ+ WY T+ L + K S
Sbjct: 940 -HLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFK----NSQVT 994
Query: 1108 LLLYVDDVLLTGNDLHEIQLVKESLHAQF--RIKDMGEAK-----FFLGLEIARSKAGIV 1160
+ L+VDD++L DL+ + + +L Q+ +I ++GE+ LGLEI + +
Sbjct: 995 ICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYM 1054
Query: 1161 -------LNQRKYALEL-LSDSGLLGGKPTTTPMDSSQ-KFGLSTDTPLSDISSYRRLIG 1211
L ++ L + L+ G P + Q + + D + ++LIG
Sbjct: 1055 KLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIG 1114
Query: 1212 KLLYL-TTTRPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLFY----PADS 1266
Y+ R D+ Y +N L+Q + P+ + +++++ L + P +
Sbjct: 1115 LASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEP 1174
Query: 1267 STTLTAFSDSDWAGCLDTRKSITGYCMFLGSSLISWRSKKQTTTSRSSCEAEYRAMAATV 1326
L A SD+ + G KS G L +I +S K + T S+ EAE A++ +V
Sbjct: 1175 DNKLVAISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESV 1233
Query: 1327 CEVQWLSYLLHDLQAPPSAPVSMYCDNQSAMHIAHNPSYHERTKHIEVDCHIVREKVQQG 1386
+ LS+L+ +L P + + I N R + +R++V
Sbjct: 1234 PLLNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGN 1293
Query: 1387 LVHLLPIASSHQLADIFTKPLTPAPFRHIFSK 1418
+H+ I + +AD+ TKPL F+ + +K
Sbjct: 1294 HLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
Score = 74.3 bits (181), Expect = 2e-12
Identities = 58/211 (27%), Positives = 95/211 (44%), Gaps = 7/211 (3%)
Query: 571 FQLIHVDIWGPVSIVSLHGFSYFLTIVDDYSRFTWIYLLKS*AE--VKNLVQEFCALVAN 628
FQ +H DI+GPV + SYF++ D+ ++F W+Y L E + ++ A + N
Sbjct: 238 FQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKN 297
Query: 629 QFETAVKTIRSDNGKEF---SLPQFYATKGIVHQTSCVETPQQNSIVERKHQHILNVARA 685
QF+ +V I+ D G E+ +L +F GI + + + + ER ++ +L+ R
Sbjct: 298 QFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRT 357
Query: 686 LLFQAHLPKIFWAHAIVHAVFLINRLPSPVLDGKCPFQILHKVLPDLTNLKVFGSLCFAS 745
L + LP W AI + + N L SP K Q D++ L FG +
Sbjct: 358 QLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVIVN 416
Query: 746 TLVSHRTKFDPRAKRCVFLGFKPGTKGYIVY 776
+ +K PR L + GYI+Y
Sbjct: 417 D-HNPNSKIHPRGIPGYALHPSRNSYGYIIY 446
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 133 bits (334), Expect = 4e-30
Identities = 170/692 (24%), Positives = 303/692 (43%), Gaps = 67/692 (9%)
Query: 770 TKGYIVYDLKSNDIAISRNVVFHENMFPY--PTQPSDLNQHQS--CPLPQASFLSDEPFE 825
T+ I++ S D + N H N+ P PT S+ N +S LP + P E
Sbjct: 658 TEKRIIHRSPSIDASPPENNSSH-NIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTE 716
Query: 826 YATQSPSEALTQPNTSEPSSD--PVLDNNHRTSTRTRKQPSYLQDYHCSLIASTAVSSSS 883
+ P + L N+ + +S + D+N T+ ++K+ L+D + S ++
Sbjct: 717 FP--DPFKELPPINSHQTNSSLGGIGDSNAYTTINSKKRS--LEDNETEIKVSRDTWNTK 772
Query: 884 SSKGTSYPLSKVISYCNLAPAYHTFVMNITAVVEPKRYSEAVKHDSWRKAMDQEIEALER 943
+ + P SK +L A V +I + RY EA+ ++ K ++ IEA +
Sbjct: 773 NMRSLEPPRSK--KRIHLIAAVKA-VKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHK 829
Query: 944 NHTWIL------VDKPHDKTPIGCKWVYRIKY----KQDGTLDRYKARLVVKGYTQLEGI 993
+L D+ +D+ I K V + K+DGT +KAR V +G I
Sbjct: 830 EVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKKRDGT---HKARFVARG-----DI 881
Query: 994 DFIDTFSPVAKMTT-----LRVLLALASSYNWFLHQLDVDNAFLHAQLDEEIYMSLPQGL 1048
DT+ + T L L+LA N+++ QLD+ +A+L+A + EE+Y+ P
Sbjct: 882 QHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP-- 939
Query: 1049 HTEKPNQVCLLQKSLYGLKQASRQWYTTLCKALHTLGFSPSSADHTLYIKKGTTGSFTAL 1108
H +++ L+KSLYGLKQ+ WY T+ L + K S +
Sbjct: 940 HLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFK----NSQVTI 995
Query: 1109 LLYVDDVLLTGNDLHEIQLVKESLHAQF--RIKDMGEAK-----FFLGLEIARSKAGIV- 1160
L+VDD++L DL+ + + +L Q+ +I ++GE+ LGLEI + +
Sbjct: 996 CLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMK 1055
Query: 1161 ------LNQRKYALEL-LSDSGLLGGKPTTTPMDSSQ-KFGLSTDTPLSDISSYRRLIGK 1212
L ++ L + L+ G P + Q + + D + ++LIG
Sbjct: 1056 LGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGL 1115
Query: 1213 LLYL-TTTRPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLFY----PADSS 1267
Y+ R D+ Y +N L+Q + P+ + +++++ L + P +
Sbjct: 1116 ASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPD 1175
Query: 1268 TTLTAFSDSDWAGCLDTRKSITGYCMFLGSSLISWRSKKQTTTSRSSCEAEYRAMAATVC 1327
L A SD+ + G KS G L +I +S K + T S+ EAE A++ +V
Sbjct: 1176 NKLVAISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVP 1234
Query: 1328 EVQWLSYLLHDLQAPPSAPVSMYCDNQSAMHIAHNPSYHE-RTKHIEVDCHIVREKVQQG 1386
+ LSYL+ +L P + D++S + I + + + R + +R++V
Sbjct: 1235 LLNNLSYLIQELNKKPIIK-GLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGN 1293
Query: 1387 LVHLLPIASSHQLADIFTKPLTPAPFRHIFSK 1418
+++ I + +AD+ TKPL F+ + +K
Sbjct: 1294 NLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1325
Score = 74.3 bits (181), Expect = 2e-12
Identities = 58/211 (27%), Positives = 95/211 (44%), Gaps = 7/211 (3%)
Query: 571 FQLIHVDIWGPVSIVSLHGFSYFLTIVDDYSRFTWIYLLKS*AE--VKNLVQEFCALVAN 628
FQ +H DI+GPV + SYF++ D+ ++F W+Y L E + ++ A + N
Sbjct: 238 FQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKN 297
Query: 629 QFETAVKTIRSDNGKEF---SLPQFYATKGIVHQTSCVETPQQNSIVERKHQHILNVARA 685
QF+ +V I+ D G E+ +L +F GI + + + + ER ++ +L+ R
Sbjct: 298 QFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRT 357
Query: 686 LLFQAHLPKIFWAHAIVHAVFLINRLPSPVLDGKCPFQILHKVLPDLTNLKVFGSLCFAS 745
L + LP W AI + + N L SP K Q D++ L FG +
Sbjct: 358 QLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVIVN 416
Query: 746 TLVSHRTKFDPRAKRCVFLGFKPGTKGYIVY 776
+ +K PR L + GYI+Y
Sbjct: 417 D-HNPNSKIHPRGIPGYALHPSRNSYGYIIY 446
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 132 bits (333), Expect = 5e-30
Identities = 157/640 (24%), Positives = 278/640 (42%), Gaps = 52/640 (8%)
Query: 812 PLPQASFLSD-EPFEYATQSPSEALTQPNTSEPSSDPVLDNNHRTSTRTRKQPSYLQDYH 870
PLP + S + + + P Q N+S D D+N T+T+++K+ L+D
Sbjct: 1147 PLPDLTHQSPTDTSDVSKDIPHIHSRQTNSSLGGMD---DSNVLTTTKSKKRS--LEDNE 1201
Query: 871 CSLIASTAVSSSSSSKGTSYPLSKVISYCNLAPAYHTFVMNITAVVEPKRYSEAVKHDSW 930
+ S ++ + + P SK NL A V +I V RY EA+ ++
Sbjct: 1202 TEIEVSRDTWNNKNMRSLEPPRSK--KRINLIAAIKG-VKSIKPVRTTLRYDEAITYNKD 1258
Query: 931 RKAMDQEIEALERNHTWIL------VDKPHDKTPIGCKWVYRIKY----KQDGTLDRYKA 980
K D+ +EA + + +L +K +D+ I K V + K+DGT +KA
Sbjct: 1259 NKEKDRYVEAYHKEISQLLKMNTWDTNKYYDRNDIDPKKVINSMFIFNKKRDGT---HKA 1315
Query: 981 RLVVKGYTQLEGIDFIDTFSPVAKMTTLRVLLALASSYNWFLHQLDVDNAFLHAQLDEEI 1040
R V +G Q D S L L++A ++++ QLD+ +A+L+A + EE+
Sbjct: 1316 RFVARGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLDISSAYLYADIKEEL 1375
Query: 1041 YMSLPQGLHTEKPNQVCLLQKSLYGLKQASRQWYTTLCKALHTLGFSPSSADHTLYIKKG 1100
Y+ P H +++ L+KSLYGLKQ+ WY T+ L + K
Sbjct: 1376 YIRPPP--HLGLNDKLLRLRKSLYGLKQSGANWYETIKSYLINCCDMQEVRGWSCVFK-- 1431
Query: 1101 TTGSFTALLLYVDDVLLTGNDLHEIQLVKESLHAQF--RIKDMGEAK-----FFLGLEIA 1153
S + L+VDD++L DL+ + + +L Q+ +I ++GE+ LGLEI
Sbjct: 1432 --NSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIK 1489
Query: 1154 RSKAGIV-LNQRKYALELLSDSGL---LGGKPTTTPMD-----SSQKFGLSTDTPLSDIS 1204
++ + L K E L + GK P + + D +
Sbjct: 1490 YQRSKYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHYIDQDELEIDEDEYKEKVH 1549
Query: 1205 SYRRLIGKLLYL-TTTRPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLFY- 1262
++LIG Y+ R D+ Y +N L+Q + P+ + +++++ L +
Sbjct: 1550 EMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWH 1609
Query: 1263 ---PADSSTTLTAFSDSDWAGCLDTRKSITGYCMFLGSSLISWRSKKQTTTSRSSCEAEY 1319
P L A SD+ + G KS G L +I +S K + T S+ EAE
Sbjct: 1610 KNKPTKPDNKLVAISDASY-GNQPYYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEI 1668
Query: 1320 RAMAATVCEVQWLSYLLHDLQAPPSAPVSMYCDNQSAMHIAHNPSYHE-RTKHIEVDCHI 1378
A++ + + LS+L+ +L P + D++S + I + + + R +
Sbjct: 1669 HAVSEAIPLLNNLSHLVQELNKKPIIK-GLLTDSRSTISIIKSTNEEKFRNRFFGTKAMR 1727
Query: 1379 VREKVQQGLVHLLPIASSHQLADIFTKPLTPAPFRHIFSK 1418
+R++V +++ I + +AD+ TKPL F+ + +K
Sbjct: 1728 LRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1767
Score = 87.8 bits (216), Expect = 2e-16
Identities = 79/325 (24%), Positives = 138/325 (42%), Gaps = 17/325 (5%)
Query: 542 SKAHVCDVCPVAKQKR----MSFPLSVTQSTAIFQLIHVDIWGPVSIVSLHGFSYFLTIV 597
+ + C C + K + L +S FQ +H DI+GPV + SYF++
Sbjct: 628 ASTYQCPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFT 687
Query: 598 DDYSRFTWIYLLKS*AE--VKNLVQEFCALVANQFETAVKTIRSDNGKEF---SLPQFYA 652
D+ +RF W+Y L E + N+ A + NQF V I+ D G E+ +L +F+
Sbjct: 688 DEKTRFQWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFT 747
Query: 653 TKGIVHQTSCVETPQQNSIVERKHQHILNVARALLFQAHLPKIFWAHAIVHAVFLINRLP 712
+GI + + + + ER ++ +LN R LL + LP W A+ + + N L
Sbjct: 748 NRGITACYTTTADSRAHGVAERLNRTLLNDCRTLLHCSGLPNHLWFSAVEFSTIIRNSLV 807
Query: 713 SPVLDGKCPFQILHKVLPDLTNLKVFGSLCFASTLVSHRTKFDPRAKRCVFLGFKPGTKG 772
SP D K Q D+T + FG + + +K PR L + G
Sbjct: 808 SPKND-KSARQHAGLAGLDITTILPFGQPVIVNN-HNPDSKIHPRGIPGYALHPSRNSYG 865
Query: 773 YIVY------DLKSNDIAISRNVVFHENMFPYPTQPSDLNQHQSCPLPQASFLSDEPFEY 826
YI+Y + + + I ++ + F Y T D + ++ Q+ +E +
Sbjct: 866 YIIYLPSLKKTVDTTNYVILQDKQSKLDQFNYDTLTFDDDLNRLTAHNQSFIEQNETEQS 925
Query: 827 ATQSPSEALTQPNTSEPSSDPVLDN 851
Q+ + E +SDP++++
Sbjct: 926 YDQNTESDHDYQSEIEINSDPLVND 950
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 130 bits (328), Expect = 2e-29
Identities = 169/692 (24%), Positives = 302/692 (43%), Gaps = 67/692 (9%)
Query: 770 TKGYIVYDLKSNDIAISRNVVFHENMFPY--PTQPSDLNQHQS--CPLPQASFLSDEPFE 825
T+ I++ S D + N H N+ P PT S+ N +S LP + P E
Sbjct: 1085 TEKRIIHRSPSIDASPPENNSSH-NIVPIKTPTTVSEQNTEESIIADLPLPDLPPESPTE 1143
Query: 826 YATQSPSEALTQPNTSEPSSD--PVLDNNHRTSTRTRKQPSYLQDYHCSLIASTAVSSSS 883
+ P + L N+ + +S + D+N T+ ++K+ L+D + S ++
Sbjct: 1144 FP--DPFKELPPINSHQTNSSLGGIGDSNAYTTINSKKRS--LEDNETEIKVSRDTWNTK 1199
Query: 884 SSKGTSYPLSKVISYCNLAPAYHTFVMNITAVVEPKRYSEAVKHDSWRKAMDQEIEALER 943
+ + P SK +L A V +I + RY EA+ ++ K ++ IEA +
Sbjct: 1200 NMRSLEPPRSK--KRIHLIAAVKA-VKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHK 1256
Query: 944 NHTWIL------VDKPHDKTPIGCKWVYRIKY----KQDGTLDRYKARLVVKGYTQLEGI 993
+L D+ +D+ I K V + K+DGT +KAR V +G I
Sbjct: 1257 EVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKKRDGT---HKARFVARG-----DI 1308
Query: 994 DFIDTFSPVAKMTT-----LRVLLALASSYNWFLHQLDVDNAFLHAQLDEEIYMSLPQGL 1048
DT+ + T L L+LA N+++ QLD+ +A+L+A + EE+Y+ P
Sbjct: 1309 QHPDTYDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP-- 1366
Query: 1049 HTEKPNQVCLLQKSLYGLKQASRQWYTTLCKALHTLGFSPSSADHTLYIKKGTTGSFTAL 1108
H +++ L+KS YGLKQ+ WY T+ L + K S +
Sbjct: 1367 HLGMNDKLIRLKKSHYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFK----NSQVTI 1422
Query: 1109 LLYVDDVLLTGNDLHEIQLVKESLHAQF--RIKDMGEAK-----FFLGLEIARSKAGIV- 1160
L+VDD++L DL+ + + +L Q+ +I ++GE+ LGLEI + +
Sbjct: 1423 CLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMK 1482
Query: 1161 ------LNQRKYALEL-LSDSGLLGGKPTTTPMDSSQ-KFGLSTDTPLSDISSYRRLIGK 1212
L ++ L + L+ G P + Q + + D + ++LIG
Sbjct: 1483 LGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGL 1542
Query: 1213 LLYL-TTTRPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLFY----PADSS 1267
Y+ R D+ Y +N L+Q + P+ + +++++ L + P +
Sbjct: 1543 ASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPD 1602
Query: 1268 TTLTAFSDSDWAGCLDTRKSITGYCMFLGSSLISWRSKKQTTTSRSSCEAEYRAMAATVC 1327
L A SD+ + G KS G L +I +S K + T S+ EAE A++ +V
Sbjct: 1603 NKLVAISDASY-GNQPYYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVP 1661
Query: 1328 EVQWLSYLLHDLQAPPSAPVSMYCDNQSAMHIAHNPSYHE-RTKHIEVDCHIVREKVQQG 1386
+ LSYL+ +L P + D++S + I + + + R + +R++V
Sbjct: 1662 LLNNLSYLIQELNKKPIIK-GLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGN 1720
Query: 1387 LVHLLPIASSHQLADIFTKPLTPAPFRHIFSK 1418
+++ I + +AD+ TKPL F+ + +K
Sbjct: 1721 NLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1752
Score = 74.3 bits (181), Expect = 2e-12
Identities = 58/211 (27%), Positives = 95/211 (44%), Gaps = 7/211 (3%)
Query: 571 FQLIHVDIWGPVSIVSLHGFSYFLTIVDDYSRFTWIYLLKS*AE--VKNLVQEFCALVAN 628
FQ +H DI+GPV + SYF++ D+ ++F W+Y L E + ++ A + N
Sbjct: 665 FQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKN 724
Query: 629 QFETAVKTIRSDNGKEF---SLPQFYATKGIVHQTSCVETPQQNSIVERKHQHILNVARA 685
QF+ +V I+ D G E+ +L +F GI + + + + ER ++ +L+ R
Sbjct: 725 QFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRT 784
Query: 686 LLFQAHLPKIFWAHAIVHAVFLINRLPSPVLDGKCPFQILHKVLPDLTNLKVFGSLCFAS 745
L + LP W AI + + N L SP K Q D++ L FG +
Sbjct: 785 QLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVIVN 843
Query: 746 TLVSHRTKFDPRAKRCVFLGFKPGTKGYIVY 776
+ +K PR L + GYI+Y
Sbjct: 844 D-HNPNSKIHPRGIPGYALHPSRNSYGYIIY 873
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 129 bits (325), Expect = 4e-29
Identities = 169/706 (23%), Positives = 289/706 (39%), Gaps = 95/706 (13%)
Query: 770 TKGYIVYDLKSNDIAISRNVVFHENMFPYPTQPSDLNQHQSCPLP--QASFLSDEPFEYA 827
T+ I++ S D + S + H + P + SD +CP + S ++D P
Sbjct: 658 TEKRIIHRSPSIDTSSSESNSLHHVV---PIKTSD-----TCPKENTEESIIADLPLPDL 709
Query: 828 TQSPSEALTQPNTSEP---------SSDPVLDNNHRTSTRTRKQPSYLQDYHCSLIASTA 878
P L+ P S + D+N T+ ++K+ L+D + S
Sbjct: 710 PPEPPTELSDSFKELPPINSRQTNSSLGGIGDSNAYTTINSKKRS--LEDNETEIKVSRD 767
Query: 879 VSSSSSSKGTSYPLSKVISYCNLAPAYHTFVMNITAVVEPKRYSEAVKHDSWRKAMDQEI 938
++ + + P SK +L A V +I + RY EA+ ++ K ++ I
Sbjct: 768 TWNTKNMRSLEPPRSK--KRIHLIAAVKA-VKSIKPIRTTLRYDEAITYNKDIKEKEKYI 824
Query: 939 EALERNHTWIL------VDKPHDKTPIGCKWVYRIKY----KQDGTLDRYKARLVVKGYT 988
EA + +L DK +D+ I K V + K+DGT +KAR V +G
Sbjct: 825 EAYHKEVNQLLKMKTWDTDKYYDRKEIDPKRVINSMFIFNRKRDGT---HKARFVARG-- 879
Query: 989 QLEGIDFIDTFSPVAKMTT-----LRVLLALASSYNWFLHQLDVDNAFLHAQLDEEIYMS 1043
I DT+ + T L L+LA N+++ QLD+ +A+L+A + EE+Y+
Sbjct: 880 ---DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIR 936
Query: 1044 LPQGLHTEKPNQVCLLQKSLYGLKQASRQWYTTLCKAL-HTLGFSPSSADHTLYIKKGTT 1102
P H +++ L+KSLYGLKQ+ WY T+ L G ++
Sbjct: 937 PPP--HLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF-----E 989
Query: 1103 GSFTALLLYVDDVLLTGNDLHEIQLVKESLHAQF--RIKDMGEAK-----FFLGLEIARS 1155
S + L+VDD++L +L+ + + + L Q+ +I ++GE+ LGLEI
Sbjct: 990 NSQVTICLFVDDMVLFSKNLNSNKRIIDKLKMQYDTKIINLGESDEEIQYDILGLEIKYQ 1049
Query: 1156 ------------------KAGIVLNQRKYALELLSDSGLLGGKPTTTPMDSSQKFGLSTD 1197
K + LN + L GL Q+ L D
Sbjct: 1050 RGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGL---------YIDQQELELEED 1100
Query: 1198 TPLSDISSYRRLIGKLLYL-TTTRPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGNP 1256
+ ++LIG Y+ R D+ Y +N L+Q + P+ + ++++I
Sbjct: 1101 DYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTR 1160
Query: 1257 GCGLFY----PADSSTTLTAFSDSDWAGCLDTRKSITGYCMFLGSSLISWRSKKQTTTSR 1312
L + P + L SD+ + G KS G L +I +S K + T
Sbjct: 1161 DKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCT 1219
Query: 1313 SSCEAEYRAMAATVCEVQWLSYLLHDLQAPPSAPVSMYCDNQSAMHIAHNPSYHERTKHI 1372
S+ EAE A++ +V + LSYL+ +L P + + I N R +
Sbjct: 1220 STTEAEIHAISESVPLLNNLSYLIQELDKKPITKGLLTDSKSTISIIISNNEEKFRNRFF 1279
Query: 1373 EVDCHIVREKVQQGLVHLLPIASSHQLADIFTKPLTPAPFRHIFSK 1418
+R++V +H+ I + +AD+ TKPL F+ + +K
Sbjct: 1280 GTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
Score = 75.1 bits (183), Expect = 1e-12
Identities = 82/337 (24%), Positives = 142/337 (41%), Gaps = 25/337 (7%)
Query: 571 FQLIHVDIWGPVSIVSLHGFSYFLTIVDDYSRFTWIYLLKS*AE--VKNLVQEFCALVAN 628
FQ +H DI+GPV + SYF++ D+ ++F W+Y L E + ++ A + N
Sbjct: 238 FQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKN 297
Query: 629 QFETAVKTIRSDNGKEF---SLPQFYATKGIVHQTSCVETPQQNSIVERKHQHILNVARA 685
QF+ +V I+ D G E+ +L +F GI + + + + ER ++ +L+ R
Sbjct: 298 QFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRT 357
Query: 686 LLFQAHLPKIFWAHAIVHAVFLINRLPSPVLDGKCPFQILHKVLPDLTNLKVFGSLCFAS 745
L + LP W AI + + N L SP K Q D++ L FG +
Sbjct: 358 QLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPFGQPVIVN 416
Query: 746 TLVSHRTKFDPRAKRCVFLGFKPGTKGYIVYDLKSNDIAISRNVVFHENMFPYPTQPSDL 805
+ +K PR L + GYI+Y + + V N + S L
Sbjct: 417 D-HNPNSKIHPRGIPGYALHPSRNSYGYIIY------LPSLKKTVDTTNYVILQGKESRL 469
Query: 806 NQHQSCPLPQASFLSDEPFEYATQSPSEALTQPNTSEPSSDPVLDNNHRTSTRTRKQPSY 865
+Q + DE T S + N + S+D ++++H + P
Sbjct: 470 DQFN-----YDALTFDEDLNRLTAS-YHSFIASNEIQQSNDLNIESDHDFQSDIELHPEQ 523
Query: 866 LQDYHCSLIASTAVSSSSSSKGTSYPL-SKVISYCNL 901
L++ + S AVS + S+ +++ SK +S N+
Sbjct: 524 LRN-----VLSKAVSPTDSTPPSTHTEDSKRVSKTNI 555
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 126 bits (317), Expect = 4e-28
Identities = 110/466 (23%), Positives = 211/466 (44%), Gaps = 38/466 (8%)
Query: 978 YKARLVVKGYTQLEGIDFIDTFSPVAKMTT----LRVLLALASSYNWFLHQLDVDNAFLH 1033
YKAR+V +G TQ DT+S + + +++ L +A++ N F+ LD+++AFL+
Sbjct: 1337 YKARIVCRGDTQSP-----DTYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLY 1391
Query: 1034 AQLDEEIYMSLPQGLHTEKPNQVCLLQKSLYGLKQASRQWYTTLCKALHTLGFSPSSADH 1093
A+L+EEIY+ H V L K+LYGLKQ+ ++W L + L+ +G +S
Sbjct: 1392 AKLEEEIYIP-----HPHDRRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTP 1446
Query: 1094 TLYIKKGTTGSFTALLLYVDDVLLTGNDLHEIQLVKESLHAQFRIKDMGEA------KFF 1147
LY T + +YVDD ++ ++ + L + F +K G
Sbjct: 1447 GLY---QTEDKNLMIAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDI 1503
Query: 1148 LGLEIARSKA----GIVLNQRKYALELLSDSGLLGGKPTTTPMDSSQKFGLSTDTPLSDI 1203
LG+++ +K + L ++ + L + ++ P S+ K D
Sbjct: 1504 LGMDLVYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSE 1563
Query: 1204 SSYRR-------LIGKLLYLT-TTRPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGN 1255
+R+ L+G+L Y+ R DI + V ++++ ++ P +++++Y+
Sbjct: 1564 EEFRQGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRY 1623
Query: 1256 PGCGLFYPADSSTT--LTAFSDSDWAGCLDTRKSITGYCMFLGSSLISWRSKKQTTTSRS 1313
G+ Y D + + A +D+ D + I G ++ G ++ + S K T S
Sbjct: 1624 KDIGIHYDRDCNKDKKVIAITDASVGSEYDAQSRI-GVILWYGMNIFNVYSNKSTNRCVS 1682
Query: 1314 SCEAEYRAMAATVCEVQWLSYLLHDLQAPPSAPVSMYCDNQSAMHIAHNPSYHERTKHIE 1373
S EAE A+ + + L L +L + + M D++ A+ + + K
Sbjct: 1683 STEAELHAIYEGYADSETLKVTLKELGEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTW 1742
Query: 1374 VDCHIVREKVQQGLVHLLPIASSHQLADIFTKPLTPAPFRHIFSKL 1419
+ I++EK+++ + LL I +AD+ TKP++ + F+ L
Sbjct: 1743 IKTEIIKEKIKEKSIKLLKITGKGNIADLLTKPVSASDFKRFIQVL 1788
Score = 69.7 bits (169), Expect = 5e-11
Identities = 72/287 (25%), Positives = 120/287 (41%), Gaps = 18/287 (6%)
Query: 518 HYRLGHPSL------VKGQSINELFPYVQCSKAHVCDVCPVAKQ-KRMSFPLSVTQSTAI 570
H R+GH + +K E ++ C C ++K KR + S+ +
Sbjct: 562 HKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTD 621
Query: 571 FQLIH---VDIWGPVSIVSLHGFSYFLTIVDDYSRF--TWIYLLKS*AEVKNLVQEFCAL 625
+ +DI+GPVS + Y L +VD+ +R+ T + K+ + V++
Sbjct: 622 HEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQY 681
Query: 626 VANQFETAVKTIRSDNGKEFS---LPQFYATKGIVHQTSCVETPQQNSIVERKHQHILNV 682
V QF+ V+ I SD G EF+ + +++ +KGI H + + N ER + I+
Sbjct: 682 VETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITD 741
Query: 683 ARALLFQAHLPKIFWAHAIVHAVFLINRLPSPVLDGKCPFQILHKVLPDLTNLKVFGSLC 742
A LL Q++L FW +A+ A + N L GK P + + + P L F
Sbjct: 742 ATTLLRQSNLRVKFWEYAVTSATNIRNYLEHK-STGKLPLKAISR-QPVTVRLMSFLPFG 799
Query: 743 FASTLVSH-RTKFDPRAKRCVFLGFKPGTKGYIVYDLKSNDIAISRN 788
+ +H K P + L P + GY + N I S N
Sbjct: 800 EKGIIWNHNHKKLKPSGLPSIILCKDPNSYGYKFFIPSKNKIVTSDN 846
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 112 bits (280), Expect = 7e-24
Identities = 58/126 (46%), Positives = 78/126 (61%), Gaps = 7/126 (5%)
Query: 901 LAPAYHTFVMNITAVVEPKRYSEAVKHDSWRKAMDQEIEALERNHTWILVDKPHDKTPIG 960
L P Y + + T EPK A+K W +AM +E++AL RN TWILV P ++ +G
Sbjct: 12 LNPKY-SLTITTTIKKEPKSVIFALKDPGWCQAMQEELDALSRNKTWILVPPPVNQNILG 70
Query: 961 CKWVYRIKYKQDGTLDRYKARLVVKGYTQLEGIDFIDTFSPVAKMTTLRVLLALA----- 1015
CKWV++ K DGTLDR KARLV KG+ Q EGI F++T+SPV + T+R +L +A
Sbjct: 71 CKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTILNVAQQLEV 130
Query: 1016 -SSYNW 1020
S NW
Sbjct: 131 GQSINW 136
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 97.1 bits (240), Expect = 3e-19
Identities = 44/79 (55%), Positives = 55/79 (68%)
Query: 1214 LYLTTTRPDIAYVVNQLSQFLSAPTNVHEAAAHRVLRYIKGNPGCGLFYPADSSTTLTAF 1273
+YLT TRPD+ + VN+LSQF SA A ++VL Y+KG G GLFY A S L AF
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 1274 SDSDWAGCLDTRKSITGYC 1292
+DSDWA C DTR+S+TG+C
Sbjct: 61 ADSDWASCPDTRRSVTGFC 79
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 42.0 bits (97), Expect = 0.012
Identities = 44/158 (27%), Positives = 63/158 (39%), Gaps = 13/158 (8%)
Query: 571 FQLIHVDIWGPVSIVSLHGFSYFLTIVDDYSRFTWIYLLKS*AEVKNLVQEFCALVANQF 630
F +D GP+ G+ Y L +VD + FTW+Y K+ V+ L +
Sbjct: 676 FDKFFIDYIGPLP--PSQGYLYVLVVVDGMTGFTWLYPTKA-PSTSATVKSLNVLTSIAI 732
Query: 631 ETAVKTIRSDNGKEF---SLPQFYATKGIVHQTSCVETPQQNSIVERKHQHILNVARALL 687
K I SD G F + ++ +GI + S PQ S VERK+ I + LL
Sbjct: 733 P---KVIHSDQGAAFTSSTFAEWAKERGIHLEFSTPYHPQSGSKVERKNSDIKRLLTKLL 789
Query: 688 FQAHLPKIFWAHAIVHAVFLINRLPSPVLDGKCPFQIL 725
W + +N SPVL P Q+L
Sbjct: 790 VGRPTK---WYDLLPVVQLALNNTYSPVLK-YTPHQLL 823
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 42.0 bits (97), Expect = 0.012
Identities = 25/82 (30%), Positives = 43/82 (51%), Gaps = 3/82 (3%)
Query: 679 ILNVARALLFQAHLPKIFWAHAIVHAVFLINRLPSPVLDGKCPFQILHKVLPDLTNLKVF 738
I+ R++L + LPK F A A AV +IN+ PS ++ P ++ + +P + L+ F
Sbjct: 5 IIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSYLRRF 64
Query: 739 GSLCFASTLVSHRTKFDPRAKR 760
G + + + K PRAK+
Sbjct: 65 GCVAY---IHCDEGKLKPRAKK 83
>POL_MMTVB (P03365) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 899
Score = 40.8 bits (94), Expect = 0.027
Identities = 37/130 (28%), Positives = 59/130 (44%), Gaps = 22/130 (16%)
Query: 592 YFLTIVDDYSRFTWIYLLKS*AEVKNLVQEFCALVANQFETAVKTIRSDNGKEF---SLP 648
Y VD YS FT+ ++ K+++Q A + + I++DN + S+
Sbjct: 657 YVHVTVDTYSHFTFA-TARTGEATKDVLQHLAQSFA--YMGIPQKIKTDNAPAYVSRSIQ 713
Query: 649 QFYATKGIVHQTSCVETPQQNSIVERKHQHILNVARALLFQAHLPKIFWA-------HAI 701
+F A I H T PQ +IVER HQ+I +A L K+ A H +
Sbjct: 714 EFLARWKISHVTGIPYNPQGQAIVERTHQNI---------KAQLNKLQKAGKYYTPHHLL 764
Query: 702 VHAVFLINRL 711
HA+F++N +
Sbjct: 765 AHALFVLNHV 774
>YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein
Length = 146
Score = 40.0 bits (92), Expect = 0.046
Identities = 28/76 (36%), Positives = 41/76 (53%), Gaps = 15/76 (19%)
Query: 370 WILDTGATDHICNTLSYFSSYKH------VEPIPVSLPNGIVETTTIK-GTIQITPSFIL 422
WI DTG T H+C+ S FSS+ V + S+P I+ + T+ GT+Q L
Sbjct: 79 WIFDTGCTSHMCHDRSIFSSFTRSSRKDFVRGVGGSIP--IMGSGTVNIGTVQ------L 130
Query: 423 ANVLFLPNFEFNLISV 438
+V ++P+ NLISV
Sbjct: 131 HDVSYVPDLPVNLISV 146
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 39.7 bits (91), Expect = 0.060
Identities = 38/146 (26%), Positives = 59/146 (40%), Gaps = 11/146 (7%)
Query: 547 CDVCPVAKQKRMSFP--LSVTQSTAIFQLIHVDIWGPVSIVSLHGFSYFLTIVDDYSRFT 604
C C V ++ P L + F ++D GP+ +G+ + L +VD + F
Sbjct: 858 CKQCLVTNATNLTSPPILRPVKPLKPFDKFYIDYIGPLP--PSNGYLHVLVVVDSMTGFV 915
Query: 605 WIYLLKS*AEVKNLVQEFCALVANQFETAVKTIRSDNGKEFSLPQF---YATKGIVHQTS 661
W+Y K+ V+ L + K + SD G F+ F KGI + S
Sbjct: 916 WLYPTKA-PSTSATVKALNMLTSIAIP---KVLHSDQGAAFTSSTFADWAKEKGIQLEFS 971
Query: 662 CVETPQQNSIVERKHQHILNVARALL 687
PQ + VERK+ I + LL
Sbjct: 972 TPYHPQSSGKVERKNSDIKRLLTKLL 997
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 39.3 bits (90), Expect = 0.078
Identities = 39/146 (26%), Positives = 58/146 (39%), Gaps = 11/146 (7%)
Query: 547 CDVCPVAKQKRMSFP--LSVTQSTAIFQLIHVDIWGPVSIVSLHGFSYFLTIVDDYSRFT 604
C C V ++ P L + F +D GP+ +G+ + L +VD + F
Sbjct: 860 CKQCLVTNAATLAAPPILRPERPVKPFDKFFIDYIGPLP--PSNGYLHVLVVVDSMTGFV 917
Query: 605 WIYLLKS*AEVKNLVQEFCALVANQFETAVKTIRSDNGKEFSLPQF---YATKGIVHQTS 661
W+Y K+ + + AL K I SD G F+ F KGI + S
Sbjct: 918 WLYPTKAPSTSATVK----ALNMLTSIAVPKVIHSDQGAAFTSATFADWAKNKGIQLEFS 973
Query: 662 CVETPQQNSIVERKHQHILNVARALL 687
PQ + VERK+ I + LL
Sbjct: 974 TPYHPQSSGKVERKNSDIKRLLTKLL 999
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.134 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 167,865,338
Number of Sequences: 164201
Number of extensions: 7116649
Number of successful extensions: 19073
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 18796
Number of HSP's gapped (non-prelim): 192
length of query: 1428
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1305
effective length of database: 39,777,331
effective search space: 51909416955
effective search space used: 51909416955
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0211.5


