

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
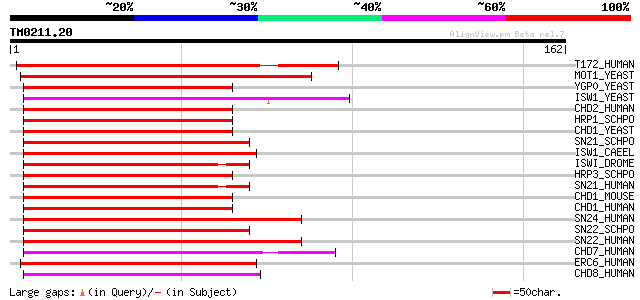
Query= TM0211.20
(162 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
T172_HUMAN (O14981) TBP-associated factor 172 (TAF-172) (TAF(II)... 100 2e-21
MOT1_YEAST (P32333) Probable helicase MOT1 93 3e-19
YGP0_YEAST (P53115) Hypothetical 171.5 kDa helicase in NUT1-ARO2... 67 2e-11
ISW1_YEAST (P38144) Chromatin remodelling complex ATPase chain I... 64 2e-10
CHD2_HUMAN (O14647) Chromodomain-helicase-DNA-binding protein 2 ... 63 3e-10
HRP1_SCHPO (Q9US25) Chromodomain helicase hrp1 62 4e-10
CHD1_YEAST (P32657) Chromo domain protein 1 62 4e-10
SN21_SCHPO (Q9UTN6) SNF2-family ATP dependent chromatin remodeli... 62 7e-10
ISW1_CAEEL (P41877) Chromatin remodelling complex ATPase chain i... 62 7e-10
ISWI_DROME (Q24368) Chromatin remodelling complex ATPase chain I... 61 1e-09
HRP3_SCHPO (O14139) Chromodomain helicase hrp3 61 1e-09
SN21_HUMAN (P28370) Possible global transcription activator SNF2... 61 1e-09
CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1 ... 61 1e-09
CHD1_HUMAN (O14646) Chromodomain-helicase-DNA-binding protein 1 ... 61 1e-09
SN24_HUMAN (P51532) Possible global transcription activator SNF2... 60 2e-09
SN22_SCHPO (O94421) SNF2-family ATP dependent chromatin remodeli... 60 2e-09
SN22_HUMAN (P51531) Possible global transcription activator SNF2... 59 4e-09
CHD7_HUMAN (Q9P2D1) Chromodomain-helicase-DNA-binding protein 7 ... 59 5e-09
ERC6_HUMAN (Q03468) DNA excision repair protein ERCC-6 (Cockayne... 58 8e-09
CHD8_HUMAN (Q9HCK8) Chromodomain-helicase-DNA-binding protein 8 ... 58 8e-09
>T172_HUMAN (O14981) TBP-associated factor 172 (TAF-172) (TAF(II)170)
Length = 1849
Score = 100 bits (248), Expect = 2e-21
Identities = 50/94 (53%), Positives = 71/94 (75%), Gaps = 5/94 (5%)
Query: 3 HVGGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMS 62
HVGG G+NLT ADT+VFVEHD N MRD QAMD+A R+GQ++VVNV+RLI R TLEE++M
Sbjct: 1707 HVGGLGLNLTGADTVVFVEHDWNPMRDLQAMDRAHRIGQKRVVNVYRLITRGTLEEKIMG 1766
Query: 63 KERFEVSVANAANNLRLLSVKSSVDHSDGVTELV 96
++F++++AN ++S ++S S G +L+
Sbjct: 1767 LQKFKMNIANT-----VISQENSSLQSMGTDQLL 1795
>MOT1_YEAST (P32333) Probable helicase MOT1
Length = 1867
Score = 92.8 bits (229), Expect = 3e-19
Identities = 43/85 (50%), Positives = 63/85 (73%)
Query: 4 VGGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSK 63
VGG G+NLT ADT++FVEHD N M D QAMD+A R+GQ+KVVNV+R+I + TLEE++M
Sbjct: 1710 VGGLGLNLTGADTVIFVEHDWNPMNDLQAMDRAHRIGQKKVVNVYRIITKGTLEEKIMGL 1769
Query: 64 ERFEVSVANAANNLRLLSVKSSVDH 88
++F++++A+ N + + S H
Sbjct: 1770 QKFKMNIASTVVNQQNSGLASMDTH 1794
>YGP0_YEAST (P53115) Hypothetical 171.5 kDa helicase in NUT1-ARO2
intergenic region
Length = 1489
Score = 66.6 bits (161), Expect = 2e-11
Identities = 30/61 (49%), Positives = 45/61 (73%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NLT+ADT++F + D N D QAMD+A RLGQ + V V+RL++R T+EE++ +
Sbjct: 1377 GGLGINLTAADTVIFYDSDWNPTIDSQAMDRAHRLGQTRQVTVYRLLVRGTIEERMRDRA 1436
Query: 65 R 65
+
Sbjct: 1437 K 1437
>ISW1_YEAST (P38144) Chromatin remodelling complex ATPase chain ISW1
(EC 3.6.1.-)
Length = 1129
Score = 63.5 bits (153), Expect = 2e-10
Identities = 33/98 (33%), Positives = 55/98 (55%), Gaps = 3/98 (3%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NLTSAD +V + D N D QAMD+A R+GQ+K V V RL+ +++EE+++ +
Sbjct: 581 GGLGINLTSADVVVLYDSDWNPQADLQAMDRAHRIGQKKQVKVFRLVTDNSVEEKILERA 640
Query: 65 RFEVSVANAA---NNLRLLSVKSSVDHSDGVTELVGSG 99
++ + N L ++ D D + ++ G
Sbjct: 641 TQKLRLDQLVIQQNRTSLKKKENKADSKDALLSMIQHG 678
>CHD2_HUMAN (O14647) Chromodomain-helicase-DNA-binding protein 2
(CHD-2)
Length = 1739
Score = 62.8 bits (151), Expect = 3e-10
Identities = 27/61 (44%), Positives = 43/61 (70%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL SADT+V + D N D QA +A R+GQ+K VN++RL+ + T+EE+++ +
Sbjct: 870 GGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGTVEEEIIERA 929
Query: 65 R 65
+
Sbjct: 930 K 930
>HRP1_SCHPO (Q9US25) Chromodomain helicase hrp1
Length = 1373
Score = 62.4 bits (150), Expect = 4e-10
Identities = 27/61 (44%), Positives = 42/61 (68%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT++ + D N D QAM +A R+GQ+ VNV+R + +DT+EE ++ +
Sbjct: 783 GGLGINLNTADTVIIFDSDWNPQADLQAMARAHRIGQKNHVNVYRFLSKDTVEEDILERA 842
Query: 65 R 65
R
Sbjct: 843 R 843
>CHD1_YEAST (P32657) Chromo domain protein 1
Length = 1468
Score = 62.4 bits (150), Expect = 4e-10
Identities = 29/61 (47%), Positives = 43/61 (69%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT+V + D N D QAM +A R+GQ+ V V+RL+ +DT+EE+V+ +
Sbjct: 774 GGLGINLMTADTVVIFDSDWNPQADLQAMARAHRIGQKNHVMVYRLVSKDTVEEEVLERA 833
Query: 65 R 65
R
Sbjct: 834 R 834
>SN21_SCHPO (Q9UTN6) SNF2-family ATP dependent chromatin remodeling
factor snf21
Length = 1199
Score = 61.6 bits (148), Expect = 7e-10
Identities = 25/66 (37%), Positives = 46/66 (68%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT++ + D N +D QA D+A R+GQ K V ++RLI ++EE ++++
Sbjct: 815 GGLGLNLQTADTVIIFDSDWNPHQDLQAQDRAHRIGQTKEVRIYRLITEKSVEENILARA 874
Query: 65 RFEVSV 70
++++ +
Sbjct: 875 QYKLDI 880
>ISW1_CAEEL (P41877) Chromatin remodelling complex ATPase chain
isw-1 (EC 3.6.1.-)
Length = 1009
Score = 61.6 bits (148), Expect = 7e-10
Identities = 28/68 (41%), Positives = 45/68 (66%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +AD ++ + D N D QAMD+A R+GQ+K V V RLI +T++E+++ K
Sbjct: 514 GGLGINLATADVVIIYDSDWNPQSDLQAMDRAHRIGQKKQVRVFRLITENTVDERIIEKA 573
Query: 65 RFEVSVAN 72
++ + N
Sbjct: 574 EAKLRLDN 581
>ISWI_DROME (Q24368) Chromatin remodelling complex ATPase chain Iswi
(EC 3.6.1.-) (Imitation swi protein) (Nucleosome
remodeling factor 140 kDa subunit) (NURF-140) (CHRAC 140
kDa subunit)
Length = 1027
Score = 61.2 bits (147), Expect = 1e-09
Identities = 31/66 (46%), Positives = 44/66 (65%), Gaps = 2/66 (3%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +AD ++ + D N D QAMD+A R+GQ+K V V RLI T+EE+++ E
Sbjct: 510 GGLGINLATADVVIIYDSDWNPQMDLQAMDRAHRIGQKKQVRVFRLITESTVEEKIV--E 567
Query: 65 RFEVSV 70
R EV +
Sbjct: 568 RAEVKL 573
>HRP3_SCHPO (O14139) Chromodomain helicase hrp3
Length = 1388
Score = 61.2 bits (147), Expect = 1e-09
Identities = 28/61 (45%), Positives = 42/61 (67%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT++ + D N D QAM +A R+GQ+ V V+RL+ +DT+EE V+ +
Sbjct: 769 GGLGINLMTADTVIIFDSDWNPQADLQAMARAHRIGQKNHVMVYRLLSKDTIEEDVLERA 828
Query: 65 R 65
R
Sbjct: 829 R 829
>SN21_HUMAN (P28370) Possible global transcription activator SNF2L1
(SWI/SNF related matrix associated actin dependent
regulator of chromatin subfamily A member 1)
Length = 976
Score = 60.8 bits (146), Expect = 1e-09
Identities = 31/66 (46%), Positives = 45/66 (67%), Gaps = 2/66 (3%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL SAD ++ + D N D QAMD+A R+GQ+K V V RLI +T+EE+++ E
Sbjct: 499 GGLGINLASADVVILYDSDWNPQVDLQAMDRAHRIGQKKPVRVFRLITDNTVEERIV--E 556
Query: 65 RFEVSV 70
R E+ +
Sbjct: 557 RAEIKL 562
>CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1
(CHD-1)
Length = 1711
Score = 60.8 bits (146), Expect = 1e-09
Identities = 26/61 (42%), Positives = 42/61 (68%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL SADT+V + D N D QA +A R+GQ+K VN++RL+ + ++EE ++ +
Sbjct: 865 GGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDILERA 924
Query: 65 R 65
+
Sbjct: 925 K 925
>CHD1_HUMAN (O14646) Chromodomain-helicase-DNA-binding protein 1
(CHD-1)
Length = 1709
Score = 60.8 bits (146), Expect = 1e-09
Identities = 26/61 (42%), Positives = 42/61 (68%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL SADT+V + D N D QA +A R+GQ+K VN++RL+ + ++EE ++ +
Sbjct: 867 GGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDILERA 926
Query: 65 R 65
+
Sbjct: 927 K 927
>SN24_HUMAN (P51532) Possible global transcription activator SNF2L4
(SNF2-beta) (BRG-1 protein) (Mitotic growth and
transcription activator) (Brahma protein homolog 1)
(SWI/SNF related matrix associated actin dependent
regulator of chromatin subfamily A m
Length = 1647
Score = 60.1 bits (144), Expect = 2e-09
Identities = 29/81 (35%), Positives = 50/81 (60%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL SADT++ + D N +D QA D+A R+GQ+ V V RL +++EE++++
Sbjct: 1159 GGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKILAAA 1218
Query: 65 RFEVSVANAANNLRLLSVKSS 85
+++++V + KSS
Sbjct: 1219 KYKLNVDQKVIQAGMFDQKSS 1239
>SN22_SCHPO (O94421) SNF2-family ATP dependent chromatin remodeling
factor snf22
Length = 1680
Score = 60.1 bits (144), Expect = 2e-09
Identities = 26/66 (39%), Positives = 45/66 (67%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT++ + D N +D QA D+A R+GQ K V + RLI ++EE ++S+
Sbjct: 1266 GGLGLNLQTADTVIIFDTDWNPHQDLQAQDRAHRIGQTKEVRILRLITEKSIEENILSRA 1325
Query: 65 RFEVSV 70
++++ +
Sbjct: 1326 QYKLDL 1331
>SN22_HUMAN (P51531) Possible global transcription activator SNF2L2
(SNF2-alpha) (SWI/SNF related matrix associated actin
dependent regulator of chromatin subfamily A member 2)
Length = 1586
Score = 59.3 bits (142), Expect = 4e-09
Identities = 29/81 (35%), Positives = 50/81 (60%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NL +ADT+V + D N +D QA D+A R+GQ+ V V RL +++EE++++
Sbjct: 1125 GGLGLNLQAADTVVIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKILAAA 1184
Query: 65 RFEVSVANAANNLRLLSVKSS 85
+++++V + KSS
Sbjct: 1185 KYKLNVDQKVIQAGMFDQKSS 1205
>CHD7_HUMAN (Q9P2D1) Chromodomain-helicase-DNA-binding protein 7
(CHD-7) (Fragment)
Length = 1967
Score = 58.9 bits (141), Expect = 5e-09
Identities = 31/91 (34%), Positives = 51/91 (55%), Gaps = 4/91 (4%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NLT+ADT + + D N D QA + R+GQ K V ++RLI R++ E ++ K
Sbjct: 607 GGLGINLTAADTCIIFDSDWNPQNDLQAQARCHRIGQSKSVKIYRLITRNSYEREMFDKA 666
Query: 65 RFEVSVANAANNLRLLSVKSSVDHSDGVTEL 95
++ + A L S+ + ++GV +L
Sbjct: 667 SLKLGLDKAV----LQSMSGRENATNGVQQL 693
>ERC6_HUMAN (Q03468) DNA excision repair protein ERCC-6 (Cockayne
syndrome protein CSB)
Length = 1493
Score = 58.2 bits (139), Expect = 8e-09
Identities = 28/69 (40%), Positives = 45/69 (64%)
Query: 4 VGGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSK 63
VGG G+NLT A+ +V + D N D QA ++A R+GQ+K V V+RL+ T+EE++ +
Sbjct: 916 VGGLGVNLTGANRVVIYDPDWNPSTDTQARERAWRIGQKKQVTVYRLLTAGTIEEKIYHR 975
Query: 64 ERFEVSVAN 72
+ F+ + N
Sbjct: 976 QIFKQFLTN 984
>CHD8_HUMAN (Q9HCK8) Chromodomain-helicase-DNA-binding protein 8
(CHD-8) (Helicase with SNF2 domain 1) (Fragment)
Length = 2004
Score = 58.2 bits (139), Expect = 8e-09
Identities = 27/69 (39%), Positives = 41/69 (59%)
Query: 5 GGFGMNLTSADTLVFVEHDRNSMRDHQAMDKARRLGQEKVVNVHRLIMRDTLEEQVMSKE 64
GG G+NLT+ADT + + D N D QA + R+GQ K V V+RLI R++ E ++ K
Sbjct: 635 GGLGINLTAADTCIIFDSDWNPQNDLQAQARCHRIGQSKAVKVYRLITRNSYEREMFDKA 694
Query: 65 RFEVSVANA 73
++ + A
Sbjct: 695 SLKLGLDKA 703
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.139 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,897,509
Number of Sequences: 164201
Number of extensions: 584663
Number of successful extensions: 2077
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 75
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1998
Number of HSP's gapped (non-prelim): 82
length of query: 162
length of database: 59,974,054
effective HSP length: 102
effective length of query: 60
effective length of database: 43,225,552
effective search space: 2593533120
effective search space used: 2593533120
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0211.20


