

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
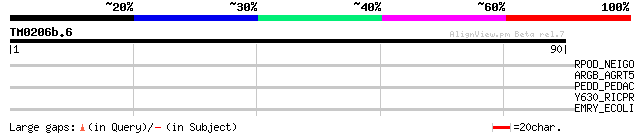
Query= TM0206b.6
(90 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RPOD_NEIGO (P52325) RNA polymerase sigma factor rpoD (Sigma-70) 28 3.1
ARGB_AGRT5 (Q8UIB7) Acetylglutamate kinase (EC 2.7.2.8) (NAG kin... 28 3.1
PEDD_PEDAC (P36497) Pediocin PA-1 transport/processing ATP-bindi... 28 4.1
Y630_RICPR (Q9ZCT3) Hypothetical UPF0118 protein RP630 27 9.1
EMRY_ECOLI (P52600) Multidrug resistance protein Y 27 9.1
>RPOD_NEIGO (P52325) RNA polymerase sigma factor rpoD (Sigma-70)
Length = 642
Score = 28.5 bits (62), Expect = 3.1
Identities = 15/41 (36%), Positives = 21/41 (50%)
Query: 25 EVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLTLCHME 65
EV ++ S L GKV N++ L++ DI L HME
Sbjct: 282 EVRFATRQIDSLSSSLRGKVENIRKLEREIRDICLDRVHME 322
>ARGB_AGRT5 (Q8UIB7) Acetylglutamate kinase (EC 2.7.2.8) (NAG
kinase) (AGK) (N-acetyl-L-glutamate
5-phosphotransferase)
Length = 294
Score = 28.5 bits (62), Expect = 3.1
Identities = 14/61 (22%), Positives = 32/61 (51%)
Query: 12 QFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLTLCHMEMLFPLS 71
+ + + + GS+ E+ A++N+ + LCGK N+ +K + ++ ++E + L
Sbjct: 96 EIVEMVLAGSINKEIVALINQTGEWAIGLCGKDGNMVFAEKAKKTVIDPDSNIERVLDLG 155
Query: 72 F 72
F
Sbjct: 156 F 156
>PEDD_PEDAC (P36497) Pediocin PA-1 transport/processing ATP-binding
protein pedD (EC 3.4.22.-) (Pediocin AcH transport
ATP-binding protein papD)
Length = 724
Score = 28.1 bits (61), Expect = 4.1
Identities = 21/78 (26%), Positives = 34/78 (42%), Gaps = 17/78 (21%)
Query: 6 FHILMEQFLPLAMWGSLP---------DEVSAVLNELSSFFRQLCGKVLNVKALDKLQHD 56
F ++++ +LP M L A++N + SFF + G+ +L D
Sbjct: 191 FQLIIDTYLPHLMTNRLSLVAIGLIVAYAFQAIINYIQSFFTIVLGQ--------RLMID 242
Query: 57 IVLTLCHMEMLFPLSFFT 74
IVL H P++FFT
Sbjct: 243 IVLKYVHHLFDLPMNFFT 260
>Y630_RICPR (Q9ZCT3) Hypothetical UPF0118 protein RP630
Length = 351
Score = 26.9 bits (58), Expect = 9.1
Identities = 24/82 (29%), Positives = 38/82 (46%), Gaps = 6/82 (7%)
Query: 10 MEQFLPLAMWGSLPDEVSAVLNELSSFFRQLCGKVLNVKALDKLQHDIVLTLCHMEMLFP 69
ME LP+ + + +SA+ N LSS+ R LN+ L + I TL +++
Sbjct: 178 MESLLPIKTRPKILEILSAINNLLSSYIR----GQLNICLLLSTYYSIAFTLIGIDLALL 233
Query: 70 LSFFTVSVHLI--LSLFFLWLL 89
L T + +I L F +LL
Sbjct: 234 LGILTGFLVIIPLLGTFISFLL 255
>EMRY_ECOLI (P52600) Multidrug resistance protein Y
Length = 512
Score = 26.9 bits (58), Expect = 9.1
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query: 13 FLPLAM--WGSLPDEVSAVLNELSSFFRQLCGKV 44
FLPL + LPD A + +S+FFR L G V
Sbjct: 383 FLPLTTISFSGLPDNKFANASSMSNFFRTLSGSV 416
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.333 0.145 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,572,737
Number of Sequences: 164201
Number of extensions: 295533
Number of successful extensions: 1274
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1271
Number of HSP's gapped (non-prelim): 5
length of query: 90
length of database: 59,974,054
effective HSP length: 66
effective length of query: 24
effective length of database: 49,136,788
effective search space: 1179282912
effective search space used: 1179282912
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0206b.6


