

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
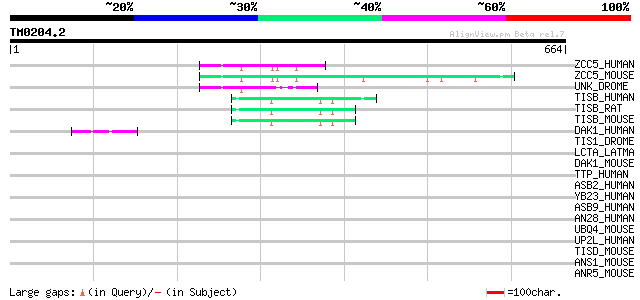
Query= TM0204.2
(664 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ZCC5_HUMAN (Q9C0B0) Zinc finger CCCH type domain containing prot... 74 9e-13
ZCC5_MOUSE (Q8BL48) Zinc finger CCCH type domain containing prot... 74 1e-12
UNK_DROME (Q86B79) Unkempt protein 68 9e-11
TISB_HUMAN (Q07352) Butyrate response factor 1 (TIS11B protein) ... 49 3e-05
TISB_RAT (P17431) Butyrate response factor 1 (TIS11B protein) (E... 48 9e-05
TISB_MOUSE (P23950) Butyrate response factor 1 (TIS11B protein) 48 9e-05
DAK1_HUMAN (P53355) Death-associated protein kinase 1 (EC 2.7.1.... 45 5e-04
TIS1_DROME (P47980) TIS11 protein (dTIS11) 43 0.003
LCTA_LATMA (Q9XZC0) Alpha-latrocrustotoxin (Alpha-LCT) (Crusta1) 43 0.003
DAK1_MOUSE (Q80YE7) Death-associated protein kinase 1 (EC 2.7.1.... 43 0.003
TTP_HUMAN (P26651) Tristetraproline (TTP) (Zinc finger protein 3... 42 0.004
ASB2_HUMAN (Q96Q27) Ankyrin repeat and SOCS box protein 2 (ASB-2) 42 0.004
YB23_HUMAN (Q9ULJ7) Hypothetical protein KIAA1223 (Fragment) 42 0.005
ASB9_HUMAN (Q96DX5) Ankyrin repeat and SOCS box protein 9 (ASB-9) 42 0.007
AN28_HUMAN (O15084) Ankyrin repeat domain protein 28 (Fragment) 42 0.007
UBQ4_MOUSE (Q99NB8) Ubiquilin 4 (Ataxin-1 ubiquitin-like interac... 41 0.009
UP2L_HUMAN (Q14157) Ubiquitin associated protein 2-like 41 0.012
TISD_MOUSE (P23949) Butyrate response factor 2 (TIS11D protein) 41 0.012
ANS1_MOUSE (P59672) Ankyrin repeat and SAM domain protein 1 40 0.015
ANR5_MOUSE (Q9D2J7) Ankyrin repeat domain protein 5 40 0.015
>ZCC5_HUMAN (Q9C0B0) Zinc finger CCCH type domain containing protein
5
Length = 810
Score = 74.3 bits (181), Expect = 9e-13
Identities = 52/172 (30%), Positives = 75/172 (43%), Gaps = 23/172 (13%)
Query: 228 SFKVKPCSRA--YSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKG-------ACQ 278
++K +PC + CP+ H ++ RRR PRK+ Y PCP + G C+
Sbjct: 215 NYKTEPCKKPPRLCRQGYACPYYHNSKD-RRRSPRKHKYRSSPCPNVKHGDEWGDPGKCE 273
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGR----KVCFFAH-----RPEELRPVYA 329
GD+C+Y H E HP Y++ C D G C FAH ++L+P A
Sbjct: 274 NGDACQYCHTRTEQQFHPEIYKSTKCNDMQQSGSCPRGPFCAFAHVEQPPLSDDLQPSSA 333
Query: 330 STGSAMPSPKSY----SSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGS 377
+ P P Y + ++V +S SP A S+L S S L GS
Sbjct: 334 VSSPTQPGPVLYMPSAAGDSVPVSPSSPHAPDLSALLCRNSSLGSPSNLCGS 385
Score = 41.6 bits (96), Expect = 0.007
Identities = 39/122 (31%), Positives = 53/122 (42%), Gaps = 22/122 (18%)
Query: 229 FKVKPCSRAYSHDWTE-----CPFVHPGENARRRDPRK----YPYSC-VPCPEFRK--GA 276
F+ + C H T+ C H RRR R+ + YS V C ++ + G
Sbjct: 40 FRTEQCPLFVQHKCTQHRPYTCFHWHFVNQRRRRSIRRRDGTFNYSPDVYCTKYDEATGL 99
Query: 277 CQKGDSCEYAH---GVFESWLHPAQYRTRLCKDET----GCGRK--VCFFAHRPEELR-P 326
C +GD C + H G E H Y+T +C ET C + C FAH P +LR P
Sbjct: 100 CPEGDECPFLHRTTGDTERRYHLRYYKTGICIHETDSKGNCTKNGLHCAFAHGPHDLRSP 159
Query: 327 VY 328
VY
Sbjct: 160 VY 161
>ZCC5_MOUSE (Q8BL48) Zinc finger CCCH type domain containing protein
5
Length = 810
Score = 73.9 bits (180), Expect = 1e-12
Identities = 102/442 (23%), Positives = 169/442 (38%), Gaps = 69/442 (15%)
Query: 228 SFKVKPCSRA--YSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKG-------ACQ 278
++K +PC + CP+ H ++ RRR PRK+ Y PCP + G C+
Sbjct: 215 NYKTEPCKKPPRLCRQGYACPYYHNSKD-RRRSPRKHKYRSSPCPNVKHGDEWGDPGKCE 273
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGR----KVCFFAH-----RPEELRPVYA 329
GD+C+Y H E HP Y++ C D G C FAH ++++P A
Sbjct: 274 NGDACQYCHTRTEQQFHPEIYKSTKCNDMQQAGSCPRGPFCAFAHIEPPPLSDDVQPSSA 333
Query: 330 STGSAMPSPKSY----SSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGNL 385
+ P P Y + ++V +S SP A S+L S L S P
Sbjct: 334 VSSPTQPGPVLYMPSAAGDSVPVSPSSPHAPDLSALLCRNSGLGSPSHLCSS-PPGPSRK 392
Query: 386 WQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEML-----------GLGSPTRQQQQQ 434
N L S PGS K R+ + E L P Q+Q
Sbjct: 393 ASNLEGLVFPGESSLAPGSYKKAPGFEREDQVGAEYLKNFKCQAKLKPHSLEPRSQEQPL 452
Query: 435 QQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNLLSQLHGLSGPSTPTHLQSQ 494
Q +Q + I + SP + +T + +N ++ + P PT +
Sbjct: 453 LQPKQDVLGILPVGSPLTSSISSSITSSLAATPPSPAGTNSTPGMNANALPFYPTSDTVE 512
Query: 495 SRHQ--------------MMQQNMNHLRSSYPSNI--------PSSPVRKPSSLGFDSS- 531
S + +++ ++ +PS++ S+PV P SLG +S
Sbjct: 513 SVIESALDDLDLNEFGVAALEKTFDNSAVPHPSSVTIGGSLLQSSAPVNIPGSLGSSASF 572
Query: 532 SAVAAAVMNSRSAAFAKRSQSFIDR------GAAGTH-HLGMSSPSNPSCR--VSSGFSD 582
+ + + S S+ F ++ Q + + G + +H LG++ ++ S FS
Sbjct: 573 HSASPSPPVSLSSHFLQQPQGHLSQSENTFLGTSASHGSLGLNGMNSSIWEHFASGSFSP 632
Query: 583 WNSPSGKLDWGVNGDELSKLRK 604
SP+ G EL++LR+
Sbjct: 633 GTSPA--FLSGPGAAELARLRQ 652
Score = 41.6 bits (96), Expect = 0.007
Identities = 39/122 (31%), Positives = 53/122 (42%), Gaps = 22/122 (18%)
Query: 229 FKVKPCSRAYSHDWTE-----CPFVHPGENARRRDPRK----YPYSC-VPCPEFRK--GA 276
F+ + C H T+ C H RRR R+ + YS V C ++ + G
Sbjct: 40 FRTEQCPLFVQHKCTQHRPYTCFHWHFVNQRRRRSIRRRDGTFNYSPDVYCTKYDEATGL 99
Query: 277 CQKGDSCEYAH---GVFESWLHPAQYRTRLCKDET----GCGRK--VCFFAHRPEELR-P 326
C +GD C + H G E H Y+T +C ET C + C FAH P +LR P
Sbjct: 100 CPEGDECPFLHRTTGDTERRYHLRYYKTGICIHETDSKGNCTKNGLHCAFAHGPHDLRSP 159
Query: 327 VY 328
VY
Sbjct: 160 VY 161
>UNK_DROME (Q86B79) Unkempt protein
Length = 599
Score = 67.8 bits (164), Expect = 9e-11
Identities = 48/154 (31%), Positives = 72/154 (46%), Gaps = 27/154 (17%)
Query: 228 SFKVKPCSRA--YSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGA-------CQ 278
++K +PC R CP H ++ +RR PRKY Y PCP + G C+
Sbjct: 194 NYKTEPCKRPPRLCRQGYACPQYHNSKD-KRRSPRKYKYRSTPCPNVKHGEEWGEPGNCE 252
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKD--ETG-CGRKV-CFFAHRPEELRPVYASTGSA 334
GD+C+Y H E HP Y++ C D + G C R V C FAH + P +
Sbjct: 253 AGDNCQYCHTRTEQQFHPEIYKSTKCNDVQQAGYCPRSVFCAFAH----VEP------CS 302
Query: 335 MPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVST 368
M P+ N++ S+ + L+ SS P++ +T
Sbjct: 303 MDDPR---ENSLSASLANTSLLTRSSAPINIPNT 333
Score = 38.1 bits (87), Expect = 0.075
Identities = 37/119 (31%), Positives = 51/119 (42%), Gaps = 21/119 (17%)
Query: 229 FKVKPCSRAYSHDWTEC-PFV----HPGENARRRDPRK----YPYSCVP-CPEFRK--GA 276
F+V+ C H + PFV H RRR RK + YS C ++ + G
Sbjct: 27 FRVEQCQSFLQHKCNQHRPFVCFNWHFQNQRRRRPVRKRDGTFNYSADNYCTKYDETTGI 86
Query: 277 CQKGDSCEYAH---GVFESWLHPAQYRTRLCKDETG----CGRK--VCFFAHRPEELRP 326
C +GD C Y H G E H Y+T +C +T C + C FAH ++ RP
Sbjct: 87 CPEGDECPYLHRTAGDTERRYHLRYYKTCMCVHDTDSRGYCVKNGLHCAFAHGMQDQRP 145
>TISB_HUMAN (Q07352) Butyrate response factor 1 (TIS11B protein)
(EGF-response factor 1) (ERF-1)
Length = 338
Score = 49.3 bits (116), Expect = 3e-05
Identities = 50/188 (26%), Positives = 73/188 (38%), Gaps = 19/188 (10%)
Query: 266 CVPCPEFRKGACQKGDSCEYAHGVFE--SWLHPAQYRTRLCKDETGCG----RKVCFFAH 319
C P E GAC+ GD C++AHG+ E S +Y+T LC+ G C F H
Sbjct: 120 CRPFEE--NGACKYGDKCQFAHGIHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIH 177
Query: 320 RPEELRPVYASTGSAMPSPK-SYSSNAVDMSVMSPLALSSSSLPMSTVSTPP----MSPL 374
EE R + + + P+ +S + + A ++ L T TPP L
Sbjct: 178 NAEERRALAGARDLSADRPRLQHSFSFAGFPSAAATAAATGLLDSPTSITPPPILSADDL 237
Query: 375 AGSLSPKSGN----LWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSPTRQ 430
GS + G + ++ +L PS+ LPG T R M SP
Sbjct: 238 LGSPTLPDGTNNPFAFSSQELASLFAPSMGLPGGGSPTTFLFRPMSESPHM--FDSPPSP 295
Query: 431 QQQQQQQQ 438
Q Q+
Sbjct: 296 QDSLSDQE 303
>TISB_RAT (P17431) Butyrate response factor 1 (TIS11B protein)
(EGF-inducible protein CMG1)
Length = 338
Score = 47.8 bits (112), Expect = 9e-05
Identities = 45/163 (27%), Positives = 66/163 (39%), Gaps = 17/163 (10%)
Query: 266 CVPCPEFRKGACQKGDSCEYAHGVFE--SWLHPAQYRTRLCKDETGCG----RKVCFFAH 319
C P E GAC+ GD C++AHG+ E S +Y+T LC+ G C F H
Sbjct: 120 CRPFEE--NGACKYGDKCQFAHGIHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIH 177
Query: 320 RPEELRPVYASTGSAMPSPK-SYSSNAVDMSVMSPLALSSSSLPMSTVSTPP----MSPL 374
EE R + + P+ +S + + A ++ L T TPP L
Sbjct: 178 NAEERRALAGGRDLSADRPRLQHSFSFAGFPSAAATAAATGLLDSPTSITPPPILSADDL 237
Query: 375 AGSLSPKSGN----LWQNKINLNLTPPSLQLPGSRLKTALSAR 413
GS + G + ++ +L PS+ LPG T R
Sbjct: 238 LGSPTLPDGTNNPFAFSSQELASLFAPSMGLPGGGSPTTFLFR 280
>TISB_MOUSE (P23950) Butyrate response factor 1 (TIS11B protein)
Length = 338
Score = 47.8 bits (112), Expect = 9e-05
Identities = 45/163 (27%), Positives = 66/163 (39%), Gaps = 17/163 (10%)
Query: 266 CVPCPEFRKGACQKGDSCEYAHGVFE--SWLHPAQYRTRLCKDETGCG----RKVCFFAH 319
C P E GAC+ GD C++AHG+ E S +Y+T LC+ G C F H
Sbjct: 120 CRPFEE--NGACKYGDKCQFAHGIHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIH 177
Query: 320 RPEELRPVYASTGSAMPSPK-SYSSNAVDMSVMSPLALSSSSLPMSTVSTPP----MSPL 374
EE R + + P+ +S + + A ++ L T TPP L
Sbjct: 178 NAEERRALAGGRDLSADRPRLQHSFSFAGFPSAAATAAATGLLDSPTSITPPPILSADDL 237
Query: 375 AGSLSPKSGN----LWQNKINLNLTPPSLQLPGSRLKTALSAR 413
GS + G + ++ +L PS+ LPG T R
Sbjct: 238 LGSPTLPDGTNNPFAFSSQELASLFAPSMGLPGGGSPTTFLFR 280
>DAK1_HUMAN (P53355) Death-associated protein kinase 1 (EC 2.7.1.37)
(DAP kinase 1)
Length = 1432
Score = 45.4 bits (106), Expect = 5e-04
Identities = 32/79 (40%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Query: 74 EKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLL 133
E TPL+ AS G +V+ L E G D+N AC D ALH AV +EV+K LL
Sbjct: 512 EGETPLLTASARGYHDIVECLAEHG-ADLN-ACDKDGHIALHLAVR---RCQMEVIKTLL 566
Query: 134 DAGSDADSLDACGNKPMNL 152
G D D GN P+++
Sbjct: 567 SQGCFVDYQDRHGNTPLHV 585
>TIS1_DROME (P47980) TIS11 protein (dTIS11)
Length = 437
Score = 42.7 bits (99), Expect = 0.003
Identities = 38/149 (25%), Positives = 58/149 (38%), Gaps = 9/149 (6%)
Query: 264 YSCVPCPEFRK-GACQKGDSCEYAHGVFES---WLHPAQYRTRLCKDETGCG----RKVC 315
Y C F + G C+ G+ C++AHG E HP +Y+T C+ G C
Sbjct: 137 YKTELCRPFEEAGECKYGEKCQFAHGSHELRNVHRHP-KYKTEYCRTFHSVGFCPYGPRC 195
Query: 316 FFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLA 375
F H +E R A+ + + S SP + SS+ ++ S+ S
Sbjct: 196 HFVHNADEARAQQAAQAAKSSTQSQSQSQQSSSQNFSPKSNQSSNQSSNSSSSSSSSGGG 255
Query: 376 GSLSPKSGNLWQNKINLNLTPPSLQLPGS 404
G N ++ L L+PP GS
Sbjct: 256 GGGGNSINNNNGSQFYLPLSPPLSMSTGS 284
>LCTA_LATMA (Q9XZC0) Alpha-latrocrustotoxin (Alpha-LCT) (Crusta1)
Length = 1395
Score = 42.7 bits (99), Expect = 0.003
Identities = 27/75 (36%), Positives = 42/75 (56%), Gaps = 4/75 (5%)
Query: 78 PLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGS 137
PL IA++ G +VK LVE ++DVN ++ T LH A S ++VVK L+ G+
Sbjct: 1095 PLFIAAMIGQYDIVKSLVEQHKIDVNTR-NKEQFTPLH---AAASNDHIDVVKYLIQKGA 1150
Query: 138 DADSLDACGNKPMNL 152
D ++ KP++L
Sbjct: 1151 DVNAKGDENLKPIDL 1165
Score = 41.2 bits (95), Expect = 0.009
Identities = 40/151 (26%), Positives = 73/151 (47%), Gaps = 22/151 (14%)
Query: 3 SGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGFWY 62
S +K + S+++V ++ +N + + L S + E +K E+G+DVN
Sbjct: 626 SAAKVLVKSNKKVKLNEMDN----NGMTPLHYASMLGNLEFVKYFTSEQGIDVNA----- 676
Query: 63 GRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKA-TALHCAVAGG 121
K + TPL +A LF V + L++ +D++ +D+A T LH A A G
Sbjct: 677 -------KTKVKNWTPLHLAILFKKFDVAQSLLQVRNIDISTR--ADQAITPLHLAAATG 727
Query: 122 SELSLEVVKVLLDAGSDADSLDACGNKPMNL 152
+ ++VK +L++G+ D A G ++L
Sbjct: 728 NS---QIVKTILNSGAVVDQETANGFTALHL 755
Score = 35.0 bits (79), Expect = 0.63
Identities = 27/89 (30%), Positives = 48/89 (53%), Gaps = 6/89 (6%)
Query: 77 TPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAG 136
TPL IAS + VK+L+E G DVN +++ T LH +A + ++ V ++ G
Sbjct: 513 TPLHIASERKNNDFVKFLLEKG-ADVNVRTFANELTPLH--LAARQDFTIIVKTLMEKRG 569
Query: 137 SDADSLDACGNKPMNLIAPAFNSSSKSRR 165
D ++ + G P++L + S+S++ R
Sbjct: 570 IDVNAKERAGFTPLHL---SITSNSRAAR 595
>DAK1_MOUSE (Q80YE7) Death-associated protein kinase 1 (EC 2.7.1.37)
(DAP kinase 1)
Length = 1442
Score = 42.7 bits (99), Expect = 0.003
Identities = 32/79 (40%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Query: 74 EKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLL 133
E TPL+ AS G +V+ L E G D+N A D ALH AV +EV+K LL
Sbjct: 510 EGETPLLTASARGYHDIVECLAEHG-ADLN-ASDKDGHIALHLAVR---RCQMEVIKTLL 564
Query: 134 DAGSDADSLDACGNKPMNL 152
GS D D GN P+++
Sbjct: 565 GHGSFVDFQDRHGNTPLHV 583
>TTP_HUMAN (P26651) Tristetraproline (TTP) (Zinc finger protein 36
homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth
factor-inducible nuclear protein NUP475) (G0/G1 switch
regulatory protein 24)
Length = 326
Score = 42.4 bits (98), Expect = 0.004
Identities = 41/134 (30%), Positives = 56/134 (41%), Gaps = 18/134 (13%)
Query: 264 YSCVPCPEFRK-GACQKGDSCEYAHGVFE---SWLHPAQYRTRLCKDETGCGR----KVC 315
Y C F + G C+ G C++AHG+ E + HP +Y+T LC GR C
Sbjct: 104 YKTELCRTFSESGRCRYGAKCQFAHGLGELRQANRHP-KYKTELCHKFYLQGRCPYGSRC 162
Query: 316 FFAHRPEE------LRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTP 369
F H P E PV + S P ++ + P SSS P S+ P
Sbjct: 163 HFIHNPSEDLAAPGHPPVLRQSISFSGLPSGRRTSPPPPGLAGPSLSSSSFSPSSSPPPP 222
Query: 370 ---PMSPLAGSLSP 380
P+SP A S +P
Sbjct: 223 GDLPLSPSAFSAAP 236
>ASB2_HUMAN (Q96Q27) Ankyrin repeat and SOCS box protein 2 (ASB-2)
Length = 587
Score = 42.4 bits (98), Expect = 0.004
Identities = 38/130 (29%), Positives = 64/130 (49%), Gaps = 13/130 (10%)
Query: 43 ALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDV 102
AL V L+V + G ++ SK + TPL +A+ G +++L + G D+
Sbjct: 193 ALHESVSRNDLEVMQILVSGGAKVESKN--AYGITPLFVAAQSGQLEALRFLAKYG-ADI 249
Query: 103 NKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSK 162
N SD A+AL+ A E EVV+ LL G+DA+ + G P+++ +S K
Sbjct: 250 NTQA-SDNASALYEACKNEHE---EVVEFLLSQGADANKTNKDGLLPLHI------ASKK 299
Query: 163 SRRKAMELLL 172
+ +++LL
Sbjct: 300 GNYRIVQMLL 309
>YB23_HUMAN (Q9ULJ7) Hypothetical protein KIAA1223 (Fragment)
Length = 768
Score = 42.0 bits (97), Expect = 0.005
Identities = 31/82 (37%), Positives = 45/82 (54%), Gaps = 5/82 (6%)
Query: 73 SEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVL 132
+EKR+ L A+ G +VV+ L+E G V V+ C ATAL A G ++VV+VL
Sbjct: 349 NEKRSALQSAAWQGHVKVVQLLIEHGAV-VDHTCNQG-ATALCIAAQEGH---IDVVQVL 403
Query: 133 LDAGSDADSLDACGNKPMNLIA 154
L+ G+D + D G M + A
Sbjct: 404 LEHGADPNHADQFGRTAMRVAA 425
Score = 37.7 bits (86), Expect = 0.097
Identities = 30/80 (37%), Positives = 42/80 (52%), Gaps = 7/80 (8%)
Query: 76 RTPLMIASLFGSTRVVKYLVETGR-VDVNKACGSDKATALHCAVAGGSELSLEVVKVLLD 134
RTPL+ A+ G VV L+ G VD + S+ T L A A G+ +EVV+ LLD
Sbjct: 121 RTPLLAAASMGHASVVNTLLFWGAAVD---SIDSEGRTVLSIASAQGN---VEVVRTLLD 174
Query: 135 AGSDADSLDACGNKPMNLIA 154
G D + D G P+++ A
Sbjct: 175 RGLDENHRDDAGWTPLHMAA 194
>ASB9_HUMAN (Q96DX5) Ankyrin repeat and SOCS box protein 9 (ASB-9)
Length = 294
Score = 41.6 bits (96), Expect = 0.007
Identities = 33/94 (35%), Positives = 47/94 (49%), Gaps = 8/94 (8%)
Query: 62 YGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGG 121
YG I K S TPL +A VK L+E+G DVN+ G D + LH
Sbjct: 156 YGGNIDHKI--SHLGTPLYLACENQQRACVKKLLESG-ADVNQGKGQD--SPLHAVARTA 210
Query: 122 SELSLEVVKVLLDAGSDADSLDACGNKPMNLIAP 155
SE E+ +L+D G+D + +A G +P+ L+ P
Sbjct: 211 SE---ELACLLMDFGADTQAKNAEGKRPVELVPP 241
>AN28_HUMAN (O15084) Ankyrin repeat domain protein 28 (Fragment)
Length = 1059
Score = 41.6 bits (96), Expect = 0.007
Identities = 31/83 (37%), Positives = 46/83 (55%), Gaps = 7/83 (8%)
Query: 73 SEKRTPLMIASLFGSTRVVKYLVETG-RVDVNKACGSDKATALHCAVAGGSELSLEVVKV 131
+EKRTPL A+ G +++ L+ +G RV+ A S T LH AVA SE E V+V
Sbjct: 45 NEKRTPLHAAAYLGDAEIIELLILSGARVN---AKDSKWLTPLHRAVASCSE---EAVQV 98
Query: 132 LLDAGSDADSLDACGNKPMNLIA 154
LL +D ++ D P+++ A
Sbjct: 99 LLKHSADVNARDKNWQTPLHIAA 121
Score = 38.1 bits (87), Expect = 0.075
Identities = 27/72 (37%), Positives = 36/72 (49%), Gaps = 7/72 (9%)
Query: 82 ASLFGSTRVVKYLVETGRVDVNKACGSDKA-TALHCAVAGGSELSLEVVKVLLDAGSDAD 140
A+ G VVK LV G C K+ T LH A + G + VVK LLD G D +
Sbjct: 186 AAYMGHIEVVKLLVSHG---AEVTCKDKKSYTPLHAAASSGM---ISVVKYLLDLGVDMN 239
Query: 141 SLDACGNKPMNL 152
+A GN P+++
Sbjct: 240 EPNAYGNTPLHV 251
Score = 37.4 bits (85), Expect = 0.13
Identities = 34/101 (33%), Positives = 52/101 (50%), Gaps = 14/101 (13%)
Query: 77 TPLMIASLFGSTRVVKYLVETGRVDVNK--ACGSDKATALHCAVAGGSELSLEVVKVLLD 134
TPL A+ G VVKYL++ G VD+N+ A G+ T LH A G ++ VV L+D
Sbjct: 214 TPLHAAASSGMISVVKYLLDLG-VDMNEPNAYGN---TPLHVACYNGQDV---VVNELID 266
Query: 135 AGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGG 175
G+ + + G P++ F ++S +ELL+ G
Sbjct: 267 CGAIVNQKNEKGFTPLH-----FAAASTHGALCLELLVGNG 302
Score = 33.9 bits (76), Expect = 1.4
Identities = 24/82 (29%), Positives = 42/82 (50%), Gaps = 1/82 (1%)
Query: 73 SEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVL 132
S +TPLM+A+ G T V+ LV + ++ S K TALH A + G E S ++
Sbjct: 860 STGKTPLMMAAENGQTNTVEMLVSSASAELTLQDNS-KNTALHLACSKGHETSALLILEK 918
Query: 133 LDAGSDADSLDACGNKPMNLIA 154
+ + ++ +A P+++ A
Sbjct: 919 ITDRNLINATNAALQTPLHVAA 940
>UBQ4_MOUSE (Q99NB8) Ubiquilin 4 (Ataxin-1 ubiquitin-like
interacting protein A1U)
Length = 596
Score = 41.2 bits (95), Expect = 0.009
Identities = 50/189 (26%), Positives = 77/189 (40%), Gaps = 16/189 (8%)
Query: 342 SSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGSLSP-KSGNLWQNK-INLNLTPPSL 399
+ + V + P S+S P ST P SP A + P SGN + +P +
Sbjct: 88 AQDPVTAAASPPSTPDSASAP----STTPASPAAAPVQPCSSGNTTSDAGSGGGPSPVAA 143
Query: 400 QLPGSRLKTALSARDFDLEMEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDL 459
+ P S + LS L + LGLGS + QQQ Q+QL+ +S + D+
Sbjct: 144 EGPSSATASILSGFGGILGLGSLGLGSANFMELQQQMQRQLMSNPEMLSQIMENPLVQDM 203
Query: 460 TPTNLDDLLGSVDSN-LLSQL--------HGLSGPSTPTHLQSQSRHQMMQQNMNHLRSS 510
+N D + + +N + QL H L+ P +R+ M Q M +
Sbjct: 204 M-SNPDLMRHMIMANPQMQQLMERNPEISHMLNNPELMRQTMELARNPAMMQEMMRNQDR 262
Query: 511 YPSNIPSSP 519
SN+ S P
Sbjct: 263 ALSNLESVP 271
>UP2L_HUMAN (Q14157) Ubiquitin associated protein 2-like
Length = 983
Score = 40.8 bits (94), Expect = 0.012
Identities = 64/248 (25%), Positives = 88/248 (34%), Gaps = 37/248 (14%)
Query: 319 HRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVSTPPMSP----- 373
H P R + + + M S AV S +P SS LP + S P MSP
Sbjct: 415 HSPFTKRQAFTPSSTMMEVFLQEKSPAVATSTAAPPP-PSSPLPSKSTSAPQMSPGSSDN 473
Query: 374 LAGSLSPKSGNLWQNKINLNLTPP----SLQLPGSR----LKTALSARDFDLEMEMLGLG 425
+ S P L Q K +LT ++++PGS L A F E +
Sbjct: 474 QSSSPQPAQQKLKQQKKKASLTSKIPALAVEMPGSADISGLNLQFGALQFGSEPVLSDYE 533
Query: 426 S-PTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNLLSQLHGLSG 484
S PT Q L A SS ++ SN + SG
Sbjct: 534 STPTTSASSSQAPSSLYTSTASESS-------------------STISSNQSQESGYQSG 574
Query: 485 PSTPTHLQSQSRHQ--MMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVMNSR 542
P T SQ+ Q + +Q R YPS+I SSP + + SS A + ++
Sbjct: 575 PIQSTTYTSQNNAQGPLYEQRSTQTR-RYPSSISSSPQKDLTQAKNGFSSVQATQLQTTQ 633
Query: 543 SAAFAKRS 550
S A S
Sbjct: 634 SVEGATGS 641
>TISD_MOUSE (P23949) Butyrate response factor 2 (TIS11D protein)
Length = 367
Score = 40.8 bits (94), Expect = 0.012
Identities = 25/84 (29%), Positives = 38/84 (44%), Gaps = 8/84 (9%)
Query: 266 CVPCPEFRKGACQKGDSCEYAHGVFE--SWLHPAQYRTRLCKDETGCG----RKVCFFAH 319
C P E G C+ G+ C++AHG E S +Y+T LC+ G C F H
Sbjct: 132 CRPFEE--SGTCKYGEKCQFAHGFHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIH 189
Query: 320 RPEELRPVYASTGSAMPSPKSYSS 343
+E RP + G A +++ +
Sbjct: 190 NADERRPAPSGGGGASGDLRAFGA 213
>ANS1_MOUSE (P59672) Ankyrin repeat and SAM domain protein 1
Length = 1150
Score = 40.4 bits (93), Expect = 0.015
Identities = 27/64 (42%), Positives = 37/64 (57%), Gaps = 5/64 (7%)
Query: 77 TPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAG 136
TPL +A+L+G VVK L+ G +C + K T LH A G + VV+VLLDAG
Sbjct: 201 TPLDLAALYGRLEVVKLLL--GAHPNLLSCSTRKHTPLHLAARNGHK---AVVQVLLDAG 255
Query: 137 SDAD 140
D++
Sbjct: 256 MDSN 259
>ANR5_MOUSE (Q9D2J7) Ankyrin repeat domain protein 5
Length = 775
Score = 40.4 bits (93), Expect = 0.015
Identities = 35/123 (28%), Positives = 62/123 (49%), Gaps = 19/123 (15%)
Query: 29 FSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGST 88
F+ + ++ + D +LK+ + E G+ V+ Y +TPLMIA G+
Sbjct: 493 FANISFITKAGDLASLKKAI-ETGIPVDMKDNTY-------------KTPLMIACASGNI 538
Query: 89 RVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGNK 148
VVK+L+E G +VN A + T LH A G + ++V++L+ AG+ D+ +
Sbjct: 539 DVVKFLIEKG-ANVN-ATDNFLWTPLHFACHAGQQ---DIVELLVKAGASIDATSINNST 593
Query: 149 PMN 151
P++
Sbjct: 594 PLS 596
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.129 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 79,916,258
Number of Sequences: 164201
Number of extensions: 3541150
Number of successful extensions: 15232
Number of sequences better than 10.0: 246
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 238
Number of HSP's that attempted gapping in prelim test: 13752
Number of HSP's gapped (non-prelim): 895
length of query: 664
length of database: 59,974,054
effective HSP length: 117
effective length of query: 547
effective length of database: 40,762,537
effective search space: 22297107739
effective search space used: 22297107739
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0204.2


