

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
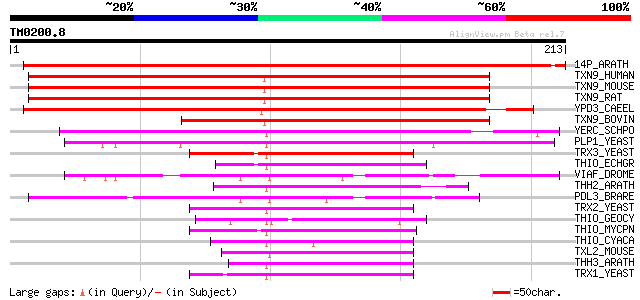
Query= TM0200.8
(213 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
14P_ARATH (O64628) UPF0071 protein At2g18990 326 3e-89
TXN9_HUMAN (O14530) Thioredoxin domain containing protein 9 (Pro... 203 2e-52
TXN9_MOUSE (Q9CQ79) Thioredoxin domain containing protein 9 (ATP... 198 7e-51
TXN9_RAT (Q8K581) Thioredoxin domain containing protein 9 (ES ce... 196 3e-50
YPD3_CAEEL (Q11183) Hypothetical UPF0071 protein C05D11.3 in chr... 176 5e-44
TXN9_BOVIN (O18883) Thioredoxin domain containing protein 9 (Pro... 146 4e-35
YERC_SCHPO (O14095) Hypothetical UPF0071 protein C2F3.12c in chr... 104 2e-22
PLP1_YEAST (Q04004) Phosducin-like protein 1 79 1e-14
TRX3_YEAST (P25372) Thioredoxin 3, mitochondrial precursor 54 2e-07
THIO_ECHGR (O17486) Thioredoxin 54 4e-07
VIAF_DROME (Q8MR62) Viral IAP-associated factor 1 homolog 49 9e-06
THH2_ARATH (Q38879) Thioredoxin H-type 2 (TRX-H-2) 48 1e-05
PDL3_BRARE (Q6P268) Phosducin-like protein 3 (Viral IAP-associat... 46 6e-05
TRX2_YEAST (P22803) Thioredoxin II (TR-II) (Thioredoxin 1) 44 2e-04
THIO_GEOCY (O96952) Thioredoxin 44 3e-04
THIO_MYCPN (P75512) Thioredoxin (TRX) 44 4e-04
THIO_CYACA (P37395) Thioredoxin 44 4e-04
TXL2_MOUSE (Q9CQM9) Thioredoxin-like protein 2 (PKC-interacting ... 43 6e-04
THH3_ARATH (Q42403) Thioredoxin H-type 3 (TRX-H-3) 43 6e-04
TRX1_YEAST (P22217) Thioredoxin I (TR-I) (Thioredoxin 2) 42 8e-04
>14P_ARATH (O64628) UPF0071 protein At2g18990
Length = 211
Score = 326 bits (835), Expect = 3e-89
Identities = 161/208 (77%), Positives = 187/208 (89%), Gaps = 1/208 (0%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
IQE++EKQVLTV +A+EDKIDDEI++L++LD DDLE LRERRL QMKKMAEK+ RWISLG
Sbjct: 5 IQEILEKQVLTVAKAMEDKIDDEIASLEKLDEDDLEVLRERRLKQMKKMAEKKKRWISLG 64
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFRENWPCKVADKHLGVLAKQHIETRFVKINAEK 125
HG+YSE+ SEKDFFSVVKAS+RVVCHF+RENWPCKV DKH+ +LAKQHIETRFVKI AEK
Sbjct: 65 HGEYSEIHSEKDFFSVVKASERVVCHFYRENWPCKVMDKHMSILAKQHIETRFVKIQAEK 124
Query: 126 SPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGE 185
SPFLAE+LKI VLPTLALIKN KVDDYVVGF+ELGG DDFSTEDLEER+A+AQVI +GE
Sbjct: 125 SPFLAERLKIVVLPTLALIKNTKVDDYVVGFNELGGKDDFSTEDLEERIARAQVIHYDGE 184
Query: 186 SSIHQARSTAQSKRSVRQATRADSSDSE 213
SS + +ST Q +R+VRQ+ R+D SDSE
Sbjct: 185 SSSLKPKSTTQVRRNVRQSARSD-SDSE 211
>TXN9_HUMAN (O14530) Thioredoxin domain containing protein 9
(Protein 1-4) (ATP binding protein associated with cell
differentiation)
Length = 226
Score = 203 bits (517), Expect = 2e-52
Identities = 90/178 (50%), Positives = 134/178 (74%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+V+E Q+L + VE+ +D EI LD++D D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 12 KVLEHQLLQTTKLVEEHLDSEIQKLDQMDEDELERLKEKRLQALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSE+DFF VK S+ VVCHF+R++ + CK+ D+HL +L+K+H+ET+F+K+N EK+
Sbjct: 72 EYREIPSERDFFQEVKESENVVCHFYRDSTFRCKILDRHLAILSKKHLETKFLKLNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L I V+PTLAL+K+ K DYVVGF +LG TDDF+TE LE RL + ++ G
Sbjct: 132 PFLCERLHIKVIPTLALLKDGKTQDYVVGFTDLGNTDDFTTETLEWRLGSSDILNYSG 189
>TXN9_MOUSE (Q9CQ79) Thioredoxin domain containing protein 9 (ATP
binding protein associated with cell differentiation)
Length = 226
Score = 198 bits (504), Expect = 7e-51
Identities = 91/178 (51%), Positives = 132/178 (74%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
EV+E Q L + VE+ +D EI LD++ D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 12 EVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+ SE+DFF VK S++VVCHF+R+ + CK+ D+HL +LAK+H+ET+F+K+N EK+
Sbjct: 72 EYREIGSERDFFQEVKESEKVVCHFYRDTTFRCKILDRHLAILAKKHLETKFLKLNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L+I V+PTLAL+++ K DYVVGF +LG TDDF+TE LE RL + VI G
Sbjct: 132 PFLCERLRIKVIPTLALLRDGKTQDYVVGFTDLGNTDDFTTETLEWRLGCSDVINYSG 189
>TXN9_RAT (Q8K581) Thioredoxin domain containing protein 9 (ES
cell-related protein)
Length = 226
Score = 196 bits (498), Expect = 3e-50
Identities = 90/178 (50%), Positives = 131/178 (73%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
EV+E Q L + VE+ +D EI LD++ D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 12 EVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+ SE+DFF VK S++VVCHF+R+ + CK+ D+HL +LAK+H+ET+F+K N EK+
Sbjct: 72 EYREIGSERDFFQEVKESEKVVCHFYRDTTFRCKILDRHLAILAKKHLETKFLKPNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L+I V+PTLAL+++ K DY+VGF +LG TDDF+TE LE RL + VI G
Sbjct: 132 PFLCERLRIKVIPTLALLRDGKTQDYIVGFTDLGNTDDFTTETLEWRLGCSDVINYSG 189
>YPD3_CAEEL (Q11183) Hypothetical UPF0071 protein C05D11.3 in
chromosome III
Length = 208
Score = 176 bits (445), Expect = 5e-44
Identities = 85/197 (43%), Positives = 131/197 (66%), Gaps = 8/197 (4%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
IQ+ +Q+L Q VE++ID E++ L+ L+ DDLE +R +R+ QMKK + R +S G
Sbjct: 5 IQQQFGEQLLRAAQVVEEQIDQEMNKLENLEEDDLEVIRRQRMEQMKKAQKDRIEMLSHG 64
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFRE-NWPCKVADKHLGVLAKQHIETRFVKINAE 124
HG Y E+ EK+FF K SD+VVC F+ N+ CK+ DKH +LA++H+ TRF+ +NAE
Sbjct: 65 HGKYEEVADEKEFFEATKKSDKVVCLFYLPGNFRCKIVDKHFEILARKHVGTRFIHVNAE 124
Query: 125 KSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
K FL +L I V+P++A++ + DY+ GFDELGG D+F+TE +E RLA+++V+ +E
Sbjct: 125 KVHFLTTRLNIRVIPSIAIVVKQQTVDYIRGFDELGGKDEFTTETMENRLARSEVLTVE- 183
Query: 185 ESSIHQARSTAQSKRSV 201
+ TA +K+ +
Sbjct: 184 ------KKHTAPAKKKI 194
>TXN9_BOVIN (O18883) Thioredoxin domain containing protein 9
(Protein 1-4) (Fragment)
Length = 179
Score = 146 bits (368), Expect = 4e-35
Identities = 65/119 (54%), Positives = 94/119 (78%), Gaps = 1/119 (0%)
Query: 67 GDYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEK 125
G+Y E+PSE+DFF VK S +VVCHF+R++ + CK+ D+HL +L+K+H+ET+F+K+N EK
Sbjct: 1 GEYREIPSERDFFQEVKESKKVVCHFYRDSTFRCKILDRHLVILSKKHLETKFLKLNVEK 60
Query: 126 SPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
+PFL E+L+I V+PTLAL+K+ K D+VVGF +LG TDDF+TE LE RL + ++ G
Sbjct: 61 APFLCERLRIKVIPTLALVKDGKTQDFVVGFSDLGNTDDFTTETLEWRLGCSDILNYSG 119
>YERC_SCHPO (O14095) Hypothetical UPF0071 protein C2F3.12c in
chromosome I
Length = 279
Score = 104 bits (259), Expect = 2e-22
Identities = 64/197 (32%), Positives = 104/197 (52%), Gaps = 13/197 (6%)
Query: 20 AVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHGDYSELPSEKDFF 79
A E + E + +LD D + RE+RL +KK + GH + + +E++
Sbjct: 88 ATESDSELEDALFSQLDEFDDTAYREQRLEMLKKEFARVEAAKEKGHMQFLTVENEREVM 147
Query: 80 SVVKASDRVVCHFFRENW-PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVL 138
+S +VV HF+ ++ CK+ D HL +AK H ET+F++I A +PFL KL + VL
Sbjct: 148 DFTLSSKKVVIHFYHPDFIRCKIIDSHLEKIAKVHWETKFIRIEAANAPFLVVKLGLKVL 207
Query: 139 PTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGESSIHQARSTAQSK 198
P + N+++ D ++GF +LG DDF T LE RL K+ S+I + + + S
Sbjct: 208 PAVLCYVNSQLVDKIIGFADLGNKDDFETSLLEFRLLKS--------SAIDRLKEESSSN 259
Query: 199 RSV----RQATRADSSD 211
+S+ Q ++D SD
Sbjct: 260 KSIYHDELQNNQSDDSD 276
>PLP1_YEAST (Q04004) Phosducin-like protein 1
Length = 230
Score = 78.6 bits (192), Expect = 1e-14
Identities = 60/194 (30%), Positives = 97/194 (49%), Gaps = 6/194 (3%)
Query: 22 EDKIDDEISALDR-LDSDD--LESLRERRLHQMKKMAEKRNRWISL-GHGDYSELPSEKD 77
E+ +D+ ++ LDR LD D L + R RL Q+ ++ + + G+G + +E D
Sbjct: 33 EENLDELLNELDRELDEDHEFLSAYRSERLQQISDHLKQVKKNVEDDGYGRLQCIDNEAD 92
Query: 78 FFSVVKASDRVVCHFFRENW-PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKIT 136
+ + VV HF E + C+ ++ L LAK+++ TRF+K+N + PFL KL I
Sbjct: 93 AIQICTKTTMVVIHFELETFGKCQYMNEKLENLAKRYLTTRFIKVNVQTCPFLVNKLNIK 152
Query: 137 VLPTLALIKNAKVDDYVVGFDELGG-TDDFSTEDLEERLAKAQVIFLEGESSIHQARSTA 195
VLP + KN VGF +LG + F LE+ LA + VI E H + +T
Sbjct: 153 VLPFVVGYKNGLEKVRYVGFSKLGNDPNGFDIRRLEQSLAHSGVIEDTFEIRKHSSVNTE 212
Query: 196 QSKRSVRQATRADS 209
+ + + +DS
Sbjct: 213 RFASTNHDRSESDS 226
>TRX3_YEAST (P25372) Thioredoxin 3, mitochondrial precursor
Length = 127
Score = 54.3 bits (129), Expect = 2e-07
Identities = 26/88 (29%), Positives = 55/88 (61%), Gaps = 3/88 (3%)
Query: 70 SELPSEKDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSP 127
++L + +F +++K +D++V F+ W PCK+ HL L + + + RFVK + ++SP
Sbjct: 28 TKLTNLTEFRNLIKQNDKLVIDFYA-TWCGPCKMMQPHLTKLIQAYPDVRFVKCDVDESP 86
Query: 128 FLAEKLKITVLPTLALIKNAKVDDYVVG 155
+A++ ++T +PT L K+ ++ ++G
Sbjct: 87 DIAKECEVTAMPTFVLGKDGQLIGKIIG 114
>THIO_ECHGR (O17486) Thioredoxin
Length = 107
Score = 53.5 bits (127), Expect = 4e-07
Identities = 29/83 (34%), Positives = 47/83 (55%), Gaps = 3/83 (3%)
Query: 80 SVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITV 137
+ +K +VC FF W PCK L +AK++ + FVK++ ++ +AEK ++T
Sbjct: 17 AAIKGDKLLVCDFFA-TWCGPCKSLAPKLDAMAKENEKVIFVKLDVDECQDVAEKYRVTA 75
Query: 138 LPTLALIKNAKVDDYVVGFDELG 160
+PTL + KN +VVG +E G
Sbjct: 76 MPTLIVFKNGCEIGHVVGANEAG 98
>VIAF_DROME (Q8MR62) Viral IAP-associated factor 1 homolog
Length = 240
Score = 48.9 bits (115), Expect = 9e-06
Identities = 57/199 (28%), Positives = 85/199 (42%), Gaps = 29/199 (14%)
Query: 22 EDKIDD-EISALDRL-DSDD---LESLRERRLHQMKKMAEKRNRWISLGHGDYSELPSEK 76
+ KIDD + LD L DS+D LE R+RR+ +M+ AEK G E+ +
Sbjct: 56 DKKIDDMSLDELDELEDSEDEAVLEQYRQRRIAEMRATAEKAR------FGSVREISGQD 109
Query: 77 DFFSVVKASDR--VVCHFFRENWP-CKVADKHLGVLAKQHIETRFVKINAEKS-PFLAEK 132
V KA + VV H + P C + H+ LA + +T+FV+ A P EK
Sbjct: 110 YVNEVTKAGEGIWVVLHLYANGVPLCALIHHHMQQLAVRFPQTKFVRSVATTCIPNFPEK 169
Query: 133 LKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGESSIHQAR 192
LPT+ + + +G EL G D + E+LE F+ G++
Sbjct: 170 ----NLPTIFIYHEGALRKQYIGPLELRG-DKLTAEELE---------FMLGQAGAVPTE 215
Query: 193 STAQSKRSVRQATRADSSD 211
T + +R AD D
Sbjct: 216 ITEDPRPQIRDKMLADLED 234
>THH2_ARATH (Q38879) Thioredoxin H-type 2 (TRX-H-2)
Length = 133
Score = 48.1 bits (113), Expect = 1e-05
Identities = 23/100 (23%), Positives = 57/100 (57%), Gaps = 11/100 (11%)
Query: 79 FSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKIT 136
F+ +K S++++ F +W PC++ + + +A + + FVK++ ++ P +A++ +T
Sbjct: 40 FNEIKESNKLLVVDFSASWCGPCRMIEPAIHAMADKFNDVDFVKLDVDELPDVAKEFNVT 99
Query: 137 VLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAK 176
+PT L+K K + ++G + ++LE++++K
Sbjct: 100 AMPTFVLVKRGKEIERIIGAKK---------DELEKKVSK 130
>PDL3_BRARE (Q6P268) Phosducin-like protein 3 (Viral IAP-associated
factor 1 homolog)
Length = 239
Score = 46.2 bits (108), Expect = 6e-05
Identities = 42/177 (23%), Positives = 92/177 (51%), Gaps = 11/177 (6%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
E + Q +VV+ ED +E+ + S++ E E ++++K++AE + + G
Sbjct: 33 EQLHLQSQSVVKTYEDMTLEELEENEDEFSEEDEHAME--MYRLKRLAEWKANQMKNVFG 90
Query: 68 DYSELPSEKDFFSVVKASDR--VVCHFFRENWP-CKVADKHLGVLAKQHIETRFVK-INA 123
+ E+ + V KA + VV H +++ P C + ++HL LA++ +++F+K I++
Sbjct: 91 ELKEISGQDYVQEVNKAGEGIWVVLHLYKQGIPLCSLINQHLAQLARKFPQSKFLKSISS 150
Query: 124 EKSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVI 180
P ++ LPTL + ++ ++ +G GG + + ++LE RL+++ +
Sbjct: 151 TCIPNYPDR----NLPTLFVYRDGEMKAQFIGPLVFGGM-NLTCDELEWRLSESGAV 202
>TRX2_YEAST (P22803) Thioredoxin II (TR-II) (Thioredoxin 1)
Length = 103
Score = 44.3 bits (103), Expect = 2e-04
Identities = 24/88 (27%), Positives = 47/88 (53%), Gaps = 2/88 (2%)
Query: 70 SELPSEKDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSP 127
++L S ++ S + + D++V F W PCK+ + A+Q+ + F K++ ++
Sbjct: 2 TQLKSASEYDSALASGDKLVVVDFFATWCGPCKMIAPMIEKFAEQYSDAAFYKLDVDEVS 61
Query: 128 FLAEKLKITVLPTLALIKNAKVDDYVVG 155
+A+K +++ +PTL K K VVG
Sbjct: 62 DVAQKAEVSSMPTLIFYKGGKEVTRVVG 89
>THIO_GEOCY (O96952) Thioredoxin
Length = 106
Score = 43.9 bits (102), Expect = 3e-04
Identities = 31/94 (32%), Positives = 53/94 (55%), Gaps = 6/94 (6%)
Query: 72 LPSEKDFFSVVK-ASDRVVCHFFRENW--PC-KVADKHLGVLAKQHIETRFVKINAEKSP 127
L ++ DF +K A D++V F +W PC ++A K++ +AK+ + F K++ +++
Sbjct: 5 LKTKADFDQALKDAGDKLVVIDFTASWCGPCQRIAPKYVE-MAKEFPDVIFYKVDVDEND 63
Query: 128 FLAEKLKITVLPTLALIKNAK-VDDYVVGFDELG 160
AE KI +PT K+ K + DYV G +E G
Sbjct: 64 ETAEAEKIQAMPTFKFYKSGKALSDYVQGANEAG 97
>THIO_MYCPN (P75512) Thioredoxin (TRX)
Length = 102
Score = 43.5 bits (101), Expect = 4e-04
Identities = 24/89 (26%), Positives = 43/89 (47%), Gaps = 3/89 (3%)
Query: 70 SELPSEKDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSP 127
+E+ S K + ++++V+ F+ E W PCK+ A + F K+N ++
Sbjct: 3 TEIKSLKQLGELFASNNKVIIDFWAE-WCGPCKITGPEFAKAASEVSTVAFAKVNVDEQT 61
Query: 128 FLAEKLKITVLPTLALIKNAKVDDYVVGF 156
+A KIT LPT+ L + + +GF
Sbjct: 62 DIAAAYKITSLPTIVLFEKGQEKHRAIGF 90
>THIO_CYACA (P37395) Thioredoxin
Length = 107
Score = 43.5 bits (101), Expect = 4e-04
Identities = 26/81 (32%), Positives = 46/81 (56%), Gaps = 3/81 (3%)
Query: 78 FFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIE-TRFVKINAEKSPFLAEKLK 134
F V S+++V F W PC++ + LA++++E + VKIN +++P ++ +
Sbjct: 12 FEKEVVNSEKLVLVDFWAPWCGPCRMISPVIDELAQEYVEQVKIVKINTDENPSISAEYG 71
Query: 135 ITVLPTLALIKNAKVDDYVVG 155
I +PTL L K+ K D V+G
Sbjct: 72 IRSIPTLMLFKDGKRVDTVIG 92
>TXL2_MOUSE (Q9CQM9) Thioredoxin-like protein 2 (PKC-interacting
cousin of thioredoxin) (PKC-theta-interacting protein)
(PKCq-interacting protein)
Length = 337
Score = 42.7 bits (99), Expect = 6e-04
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 1/75 (1%)
Query: 82 VKASDRVVCHFFRENWP-CKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPT 140
+K +V HF+ P C + + LAK+H FVK+ AE P ++EK +I+ +PT
Sbjct: 30 LKTKSLLVVHFWAPWAPQCVQMNDVMAELAKEHPHVSFVKLEAEAVPEVSEKYEISSVPT 89
Query: 141 LALIKNAKVDDYVVG 155
KN++ D + G
Sbjct: 90 FLFFKNSQKVDRLDG 104
>THH3_ARATH (Q42403) Thioredoxin H-type 3 (TRX-H-3)
Length = 118
Score = 42.7 bits (99), Expect = 6e-04
Identities = 20/73 (27%), Positives = 39/73 (53%), Gaps = 2/73 (2%)
Query: 85 SDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSPFLAEKLKITVLPTLA 142
S +++ F W PC+ LAK+H++ F K++ ++ +AE+ K+ +PT
Sbjct: 26 SKKLIVIDFTATWCPPCRFIAPVFADLAKKHLDVVFFKVDVDELNTVAEEFKVQAMPTFI 85
Query: 143 LIKNAKVDDYVVG 155
+K ++ + VVG
Sbjct: 86 FMKEGEIKETVVG 98
>TRX1_YEAST (P22217) Thioredoxin I (TR-I) (Thioredoxin 2)
Length = 102
Score = 42.4 bits (98), Expect = 8e-04
Identities = 25/88 (28%), Positives = 48/88 (54%), Gaps = 3/88 (3%)
Query: 70 SELPSEKDFFSVVKASDRVVCHFFRENW--PCKVADKHLGVLAKQHIETRFVKINAEKSP 127
++ + +F S + A D++V F W PCK+ + ++Q+ + F K++ ++
Sbjct: 2 TQFKTASEFDSAI-AQDKLVVVDFYATWCGPCKMIAPMIEKFSEQYPQADFYKLDVDELG 60
Query: 128 FLAEKLKITVLPTLALIKNAKVDDYVVG 155
+A+K +++ +PTL L KN K VVG
Sbjct: 61 DVAQKNEVSAMPTLLLFKNGKEVAKVVG 88
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,685,443
Number of Sequences: 164201
Number of extensions: 913665
Number of successful extensions: 3477
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 69
Number of HSP's successfully gapped in prelim test: 70
Number of HSP's that attempted gapping in prelim test: 3399
Number of HSP's gapped (non-prelim): 142
length of query: 213
length of database: 59,974,054
effective HSP length: 106
effective length of query: 107
effective length of database: 42,568,748
effective search space: 4554856036
effective search space used: 4554856036
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0200.8


