

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0193.1
(385 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
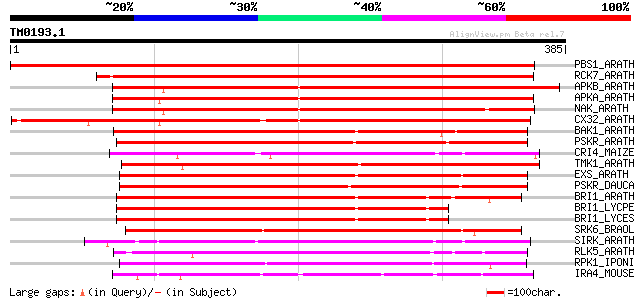
Score E
Sequences producing significant alignments: (bits) Value
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 438 e-122
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 406 e-113
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 314 3e-85
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 311 2e-84
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 308 2e-83
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 287 3e-77
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 243 5e-64
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 233 5e-61
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 233 5e-61
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 233 8e-61
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 230 5e-60
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 228 2e-59
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 226 6e-59
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 214 4e-55
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 213 7e-55
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 209 1e-53
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 199 8e-51
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 192 2e-48
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 191 4e-48
IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (... 187 3e-47
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 438 bits (1127), Expect = e-122
Identities = 222/366 (60%), Positives = 271/366 (73%), Gaps = 2/366 (0%)
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGER-KRETKGKGKGVSS 59
M CFSC S + + N + S N + + GE+ +T G K
Sbjct: 1 MGCFSCFDSSDDEKLNPVDESNHGQKKQSQPTVSNNISGLPSGGEKLSSKTNGGSKRELL 60
Query: 60 SNGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHD 118
G+ AA +F FRELA AT NF +GEGGFG+VYKGRL +TG+ VAVKQL +
Sbjct: 61 LPRDGLGQIAAHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRN 120
Query: 119 GRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKE 178
G QG +EF++EVLMLSLLHH NLV LIGYC DGDQRLLVYE+MP+GSLEDHL +L DKE
Sbjct: 121 GLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKE 180
Query: 179 PLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPV 238
L+W+ RMK+A GAA+GLE+LH A+PPVIYRD KS+NILLD F+PKLSDFGLAKLGP
Sbjct: 181 ALDWNMRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPT 240
Query: 239 GDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQN 298
GD +HVSTRVMGTYGYCAPEYAM+G+LT+KSD+YSFGVV LEL+TGR+AID+ GEQN
Sbjct: 241 GDKSHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQN 300
Query: 299 LVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVAL 358
LV+WARP F+DRR+F + DP L+GRFP+R L+QA+A+ +MC+QEQ RPLI D+V AL
Sbjct: 301 LVAWARPLFNDRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL 360
Query: 359 EYLASQ 364
YLA+Q
Sbjct: 361 SYLANQ 366
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 406 bits (1044), Expect = e-113
Identities = 200/304 (65%), Positives = 243/304 (79%), Gaps = 2/304 (0%)
Query: 61 NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTT-GEAVAVKQLSHDG 119
N GK A +F F+ELA+AT NF+ +GEGGFGKV+KG + + VA+KQL +G
Sbjct: 80 NDQVTGKKAQ-TFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNG 138
Query: 120 RQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEP 179
QG +EFV+EVL LSL H NLV+LIG+C +GDQRLLVYEYMP GSLEDHL L K+P
Sbjct: 139 VQGIREFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKP 198
Query: 180 LNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVG 239
L+W+TRMK+A GAARGLEYLH PPVIYRDLK +NILL ++ PKLSDFGLAK+GP G
Sbjct: 199 LDWNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSG 258
Query: 240 DNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNL 299
D THVSTRVMGTYGYCAP+YAM+G+LT KSDIYSFGVVLLEL+TGR+AID ++ +QNL
Sbjct: 259 DKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNL 318
Query: 300 VSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
V WARP F DRR F MVDPLLQG++P R L+QA+AI+AMC+QEQP RP+++D+V+AL
Sbjct: 319 VGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALN 378
Query: 360 YLAS 363
+LAS
Sbjct: 379 FLAS 382
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 314 bits (804), Expect = 3e-85
Identities = 161/320 (50%), Positives = 217/320 (67%), Gaps = 11/320 (3%)
Query: 72 SFGFRELADATRNFKEANLIGEGGFGKVYKGRLT----------TGEAVAVKQLSHDGRQ 121
SF F EL ATRNF+ +++GEGGFG V+KG + TG +AVK+L+ DG Q
Sbjct: 56 SFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQ 115
Query: 122 GFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLN 181
G QE++ EV L H NLV+LIGYC + + RLLVYE+MP GSLE+HLF +PL+
Sbjct: 116 GHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLS 175
Query: 182 WSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDN 241
W+ R+KVA+GAA+GL +LH A+ VIYRD K++NILLD+E+N KLSDFGLAK GP GD
Sbjct: 176 WTLRLKVALGAAKGLAFLH-NAETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDK 234
Query: 242 THVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVS 301
+HVSTR+MGTYGY APEY +G LT KSD+YS+GVVLLE+L+GRRA+D +R PGEQ LV
Sbjct: 235 SHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVE 294
Query: 302 WARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYL 361
WARP +++R+ ++D LQ ++ + + CL + K RP + ++V LE++
Sbjct: 295 WARPLLANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHLEHI 354
Query: 362 ASQGSPEAHYLYGVQQRPSR 381
+ + VQ+R R
Sbjct: 355 QTLNEAGGRNIDMVQRRMRR 374
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 311 bits (797), Expect = 2e-84
Identities = 158/302 (52%), Positives = 214/302 (70%), Gaps = 11/302 (3%)
Query: 72 SFGFRELADATRNFKEANLIGEGGFGKVYKG----------RLTTGEAVAVKQLSHDGRQ 121
SF F EL ATRNF+ +++GEGGFG V+KG R TG +AVK+L+ DG Q
Sbjct: 55 SFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQ 114
Query: 122 GFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLN 181
G QE++ EV L H +LV+LIGYC + + RLLVYE+MP GSLE+HLF +PL+
Sbjct: 115 GHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPLS 174
Query: 182 WSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDN 241
W R+KVA+GAA+GL +LH +++ VIYRD K++NILLD+E+N KLSDFGLAK GP+GD
Sbjct: 175 WKLRLKVALGAAKGLAFLH-SSETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDK 233
Query: 242 THVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVS 301
+HVSTRVMGT+GY APEY +G LT KSD+YSFGVVLLELL+GRRA+D +R GE+NLV
Sbjct: 234 SHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVE 293
Query: 302 WARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYL 361
WA+PY ++R+ ++D LQ ++ + ++ CL + K RP ++++V LE++
Sbjct: 294 WAKPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLEHI 353
Query: 362 AS 363
S
Sbjct: 354 QS 355
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 308 bits (789), Expect = 2e-83
Identities = 162/305 (53%), Positives = 218/305 (71%), Gaps = 15/305 (4%)
Query: 72 SFGFRELADATRNFKEANLIGEGGFGKVYKGRLT----------TGEAVAVKQLSHDGRQ 121
+F EL ATRNF+ +++GEGGFG V+KG + TG +AVK+L+ +G Q
Sbjct: 55 NFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQ 114
Query: 122 GFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLN 181
G +E++ E+ L L H NLV+LIGYC + + RLLVYE+M GSLE+HLF +PL+
Sbjct: 115 GHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPLS 174
Query: 182 WSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDN 241
W+TR+++A+GAARGL +LH A P VIYRD K++NILLD+ +N KLSDFGLA+ GP+GDN
Sbjct: 175 WNTRVRMALGAARGLAFLH-NAQPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMGDN 233
Query: 242 THVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVS 301
+HVSTRVMGT GY APEY +G L++KSD+YSFGVVLLELL+GRRAID ++ GE NLV
Sbjct: 234 SHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNLVD 293
Query: 302 WARPYFSDRRRFGHMVDPLLQGRFP-SRCLHQAIAITAM-CLQEQPKFRPLITDIVVALE 359
WARPY +++RR ++DP LQG++ +R L IA+ A+ C+ K RP + +IV +E
Sbjct: 294 WARPYLTNKRRLLRVMDPRLQGQYSLTRAL--KIAVLALDCISIDAKSRPTMNEIVKTME 351
Query: 360 YLASQ 364
L Q
Sbjct: 352 ELHIQ 356
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 287 bits (735), Expect = 3e-77
Identities = 151/372 (40%), Positives = 231/372 (61%), Gaps = 18/372 (4%)
Query: 2 SCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGK--GKGVSS 59
+C S F S+ + +AT + S+G + A T+ ++++ G+ S
Sbjct: 3 ACIS--FFSSSSPSKTGLHSHATTNNHSNGTEFSSTTGATTNSSVGQQSQFSDISTGIIS 60
Query: 60 SNGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKG----------RLTTGEA 109
+G + F +L AT+NFK +++G+GGFGKVY+G R+ +G
Sbjct: 61 DSGKLLESPNLKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMI 120
Query: 110 VAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDH 169
VA+K+L+ + QGF E+ EV L +L H NLV+L+GYC + + LLVYE+MP GSLE H
Sbjct: 121 VAIKRLNSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESH 180
Query: 170 LFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSD 229
LF + +P W R+K+ +GAARGL +LH + VIYRD K++NILLD+ ++ KLSD
Sbjct: 181 LFRRN---DPFPWDLRIKIVIGAARGLAFLH-SLQREVIYRDFKASNILLDSNYDAKLSD 236
Query: 230 FGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAID 289
FGLAKLGP + +HV+TR+MGTYGY APEY +G L +KSD+++FGVVLLE++TG A +
Sbjct: 237 FGLAKLGPADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHN 296
Query: 290 TSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRP 349
T R G+++LV W RP S++ R ++D ++G++ ++ + IT C++ PK RP
Sbjct: 297 TKRPRGQESLVDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRP 356
Query: 350 LITDIVVALEYL 361
+ ++V LE++
Sbjct: 357 HMKEVVEVLEHI 368
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 243 bits (621), Expect = 5e-64
Identities = 131/290 (45%), Positives = 183/290 (62%), Gaps = 5/290 (1%)
Query: 73 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQ-EFVMEVL 131
F REL A+ NF N++G GGFGKVYKGRL G VAVK+L + QG + +F EV
Sbjct: 277 FSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEVE 336
Query: 132 MLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVG 191
M+S+ H NL+RL G+C +RLLVY YM GS+ L E + PL+W R ++A+G
Sbjct: 337 MISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALG 396
Query: 192 AARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGT 251
+ARGL YLH DP +I+RD+K+ANILLD EF + DFGLAKL D THV+T V GT
Sbjct: 397 SARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKD-THVTTAVRGT 455
Query: 252 YGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQN--LVSWARPYFSD 309
G+ APEY +GK + K+D++ +GV+LLEL+TG+RA D +R + + L+ W + +
Sbjct: 456 IGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKE 515
Query: 310 RRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
++ +VD LQG + + Q I + +C Q P RP ++++V LE
Sbjct: 516 -KKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLE 564
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 233 bits (595), Expect = 5e-61
Identities = 129/286 (45%), Positives = 175/286 (61%), Gaps = 4/286 (1%)
Query: 75 FRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLS 134
+ +L D+T +F +AN+IG GGFG VYK L G+ VA+K+LS D Q +EF EV LS
Sbjct: 724 YDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLS 783
Query: 135 LLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAAR 194
H NLV L G+C + RLL+Y YM GSL+ L E + L W TR+++A GAA+
Sbjct: 784 RAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAK 843
Query: 195 GLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKL-GPVGDNTHVSTRVMGTYG 253
GL YLH DP +++RD+KS+NILLD FN L+DFGLA+L P THVST ++GT G
Sbjct: 844 GLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPY--ETHVSTDLVGTLG 901
Query: 254 YCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRF 313
Y PEY + T K D+YSFGVVLLELLT +R +D + G ++L+SW R
Sbjct: 902 YIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWV-VKMKHESRA 960
Query: 314 GHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
+ DPL+ + + + + + I +CL E PK RP +V L+
Sbjct: 961 SEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWLD 1006
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 233 bits (595), Expect = 5e-61
Identities = 135/307 (43%), Positives = 186/307 (59%), Gaps = 16/307 (5%)
Query: 70 AASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQL--SHDGRQGFQEFV 127
A F + EL AT F E + +G+G F V+KG L G VAVK+ + D ++ +EF
Sbjct: 490 AQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFH 549
Query: 128 MEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEP-----LNW 182
E+ +LS L+H +L+ L+GYC DG +RLLVYE+M GSL HL H K+P LNW
Sbjct: 550 NELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHL----HGKDPNLKKRLNW 605
Query: 183 STRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNT 242
+ R+ +AV AARG+EYLH A PPVI+RD+KS+NIL+D + N +++DFGL+ LGP T
Sbjct: 606 ARRVTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGT 665
Query: 243 HVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSW 302
+S GT GY PEY LT KSD+YSFGVVLLE+L+GR+AID G N+V W
Sbjct: 666 PLSELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEEG--NIVEW 723
Query: 303 ARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLA 362
A P F ++DP+L L + ++ C++ + K RP + + ALE+
Sbjct: 724 AVPLIKAGDIFA-ILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHAL 782
Query: 363 S--QGSP 367
+ GSP
Sbjct: 783 ALLMGSP 789
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 233 bits (593), Expect = 8e-61
Identities = 124/295 (42%), Positives = 178/295 (60%), Gaps = 6/295 (2%)
Query: 78 LADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHD--GRQGFQEFVMEVLMLSL 135
L T NF N++G GGFG VYKG L G +AVK++ + +GF EF E+ +L+
Sbjct: 581 LRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTK 640
Query: 136 LHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDK-EPLNWSTRMKVAVGAAR 194
+ H +LV L+GYC DG+++LLVYEYMP G+L HLFE S + +PL W R+ +A+ AR
Sbjct: 641 VRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDVAR 700
Query: 195 GLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGY 254
G+EYLH A I+RDLK +NILL ++ K++DFGL +L P G + + TR+ GT+GY
Sbjct: 701 GVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGS-IETRIAGTFGY 759
Query: 255 CAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSW-ARPYFSDRRRF 313
APEYA++G++T K D+YSFGV+L+EL+TGR+++D S+ +LVSW R Y + F
Sbjct: 760 LAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYINKEASF 819
Query: 314 GHMVDPLLQ-GRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQGSP 367
+D + +H + C +P RP + V L L P
Sbjct: 820 KKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNILSSLVELWKP 874
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 230 bits (586), Expect = 5e-60
Identities = 126/284 (44%), Positives = 180/284 (63%), Gaps = 3/284 (1%)
Query: 77 ELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLL 136
++ +AT +F + N+IG+GGFG VYK L + VAVK+LS QG +EF+ E+ L +
Sbjct: 909 DIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKV 968
Query: 137 HHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGL 196
H NLV L+GYC+ +++LLVYEYM GSL+ L + E L+WS R+K+AVGAARGL
Sbjct: 969 KHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARGL 1028
Query: 197 EYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCA 256
+LH P +I+RD+K++NILLD +F PK++DFGLA+L + +HVST + GT+GY
Sbjct: 1029 AFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACE-SHVSTVIAGTFGYIP 1087
Query: 257 PEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGE-QNLVSWARPYFSDRRRFGH 315
PEY S + T K D+YSFGV+LLEL+TG+ + E NLV WA + +
Sbjct: 1088 PEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKAV-D 1146
Query: 316 MVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
++DPLL + + I +CL E P RP + D++ AL+
Sbjct: 1147 VIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKALK 1190
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 228 bits (582), Expect = 2e-59
Identities = 124/283 (43%), Positives = 174/283 (60%), Gaps = 2/283 (0%)
Query: 77 ELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLL 136
++ +T +F +AN+IG GGFG VYK L G VA+K+LS D Q +EF EV LS
Sbjct: 735 DILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVETLSRA 794
Query: 137 HHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGL 196
H NLV L+GYC + +LL+Y YM GSL+ L E L+W TR+++A GAA GL
Sbjct: 795 QHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAEGL 854
Query: 197 EYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCA 256
YLH + +P +++RD+KS+NILL + F L+DFGLA+L + +THV+T ++GT GY
Sbjct: 855 AYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARL-ILPYDTHVTTDLVGTLGYIP 913
Query: 257 PEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHM 316
PEY + T K D+YSFGVVLLELLTGRR +D + G ++L+SW +++R +
Sbjct: 914 PEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRE-SEI 972
Query: 317 VDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
DP + + + + + I CL E PK RP +V LE
Sbjct: 973 FDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLE 1015
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 226 bits (577), Expect = 6e-59
Identities = 126/284 (44%), Positives = 173/284 (60%), Gaps = 7/284 (2%)
Query: 75 FRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLS 134
F +L AT F +LIG GGFG VYK L G AVA+K+L H QG +EF+ E+ +
Sbjct: 873 FADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEMETIG 932
Query: 135 LLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAAR 194
+ H NLV L+GYC GD+RLLVYE+M GSLED L + LNWSTR K+A+G+AR
Sbjct: 933 KIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSAR 992
Query: 195 GLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVM-GTYG 253
GL +LH P +I+RD+KS+N+LLD ++SDFG+A+L D TH+S + GT G
Sbjct: 993 GLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLSVSTLAGTPG 1051
Query: 254 YCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRF 313
Y PEY S + + K D+YS+GVVLLELLTG+R D S G+ NLV W + + + R
Sbjct: 1052 YVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD-SPDFGDNNLVGWVKQH--AKLRI 1108
Query: 314 GHMVDPLLQGRFPSRCLH--QAIAITAMCLQEQPKFRPLITDIV 355
+ DP L P+ + Q + + CL ++ RP + ++
Sbjct: 1109 SDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVM 1152
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 214 bits (544), Expect = 4e-55
Identities = 112/231 (48%), Positives = 149/231 (64%), Gaps = 3/231 (1%)
Query: 75 FRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLS 134
F +L +AT F +L+G GGFG VYK +L G VA+K+L H QG +EF E+ +
Sbjct: 878 FADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETIG 937
Query: 135 LLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAAR 194
+ H NLV L+GYC G++RLLVYEYM GSLED L + LNW R K+A+GAAR
Sbjct: 938 KIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAIGAAR 997
Query: 195 GLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVM-GTYG 253
GL +LH P +I+RD+KS+N+LLD ++SDFG+A+L D TH+S + GT G
Sbjct: 998 GLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLSVSTLAGTPG 1056
Query: 254 YCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWAR 304
Y PEY S + + K D+YS+GVVLLELLTG++ D S G+ NLV W +
Sbjct: 1057 YVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGWVK 1106
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 213 bits (542), Expect = 7e-55
Identities = 112/231 (48%), Positives = 149/231 (64%), Gaps = 3/231 (1%)
Query: 75 FRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLS 134
F +L +AT F +L+G GGFG VYK +L G VA+K+L H QG +EF E+ +
Sbjct: 878 FADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETIG 937
Query: 135 LLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAAR 194
+ H NLV L+GYC G++RLLVYEYM GSLED L + LNW R K+A+GAAR
Sbjct: 938 KIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAIGAAR 997
Query: 195 GLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVM-GTYG 253
GL +LH P +I+RD+KS+N+LLD ++SDFG+A+L D TH+S + GT G
Sbjct: 998 GLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMD-THLSVSTLAGTPG 1056
Query: 254 YCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWAR 304
Y PEY S + + K D+YS+GVVLLELLTG++ D S G+ NLV W +
Sbjct: 1057 YVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGWVK 1106
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 209 bits (532), Expect = 1e-53
Identities = 112/282 (39%), Positives = 176/282 (61%), Gaps = 9/282 (3%)
Query: 81 ATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTN 140
AT NF N +G+GGFG VYKGRL G+ +AVK+LS QG EF+ EV +++ L H N
Sbjct: 524 ATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMNEVTLIARLQHIN 583
Query: 141 LVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLH 200
LV+++G C +GD+++L+YEY+ SL+ +LF + + LNW+ R + G ARGL YLH
Sbjct: 584 LVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTR-RSKLNWNERFDITNGVARGLLYLH 642
Query: 201 CTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYA 260
+ +I+RDLK +NILLD PK+SDFG+A++ + + +V+GTYGY +PEYA
Sbjct: 643 QDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGTYGYMSPEYA 702
Query: 261 MSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPL 320
M G + KSD++SFGV++LE+++G++ E +L+S+ + + R +VDP+
Sbjct: 703 MYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGRAL-EIVDPV 761
Query: 321 L-------QGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIV 355
+ F + + + I I +C+QE + RP ++ +V
Sbjct: 762 IVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVV 803
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 199 bits (507), Expect = 8e-51
Identities = 119/311 (38%), Positives = 183/311 (58%), Gaps = 9/311 (2%)
Query: 53 KGKGVSSSNGSSNG--KTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAV 110
K K + G NG KTA F + E+ + T NF+ +IG+GGFGKVY G + GE V
Sbjct: 542 KKKQQRGTLGERNGPLKTAKRYFKYSEVVNITNNFER--VIGKGGFGKVYHG-VINGEQV 598
Query: 111 AVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHL 170
AVK LS + QG++EF EV +L +HHTNL L+GYC + + +L+YEYM +L D+L
Sbjct: 599 AVKVLSEESAQGYKEFRAEVDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYL 658
Query: 171 FELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDF 230
L+W R+K+++ AA+GLEYLH PP+++RD+K NILL+ + K++DF
Sbjct: 659 --AGKRSFILSWEERLKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADF 716
Query: 231 GLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDT 290
GL++ V + +ST V G+ GY PEY + ++ KSD+YS GVVLLE++TG+ AI +
Sbjct: 717 GLSRSFSVEGSGQISTVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIAS 776
Query: 291 SRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPL 350
S+ + ++ R ++ G +VD L+ R+ + I C + RP
Sbjct: 777 SKTE-KVHISDHVRSILANGDIRG-IVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPT 834
Query: 351 ITDIVVALEYL 361
++ +V+ L+ +
Sbjct: 835 MSQVVMELKQI 845
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 192 bits (487), Expect = 2e-48
Identities = 118/299 (39%), Positives = 170/299 (56%), Gaps = 19/299 (6%)
Query: 73 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQE------- 125
F E+AD E N+IG G GKVYK L GE VAVK+L+ + G E
Sbjct: 674 FSEHEIADC---LDEKNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLN 730
Query: 126 ---FVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNW 182
F EV L + H ++VRL C+ GD +LLVYEYMP GSL D L L W
Sbjct: 731 RDVFAAEVETLGTIRHKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGW 790
Query: 183 STRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPV-GDN 241
R+++A+ AA GL YLH PP+++RD+KS+NILLD+++ K++DFG+AK+G + G
Sbjct: 791 PERLRIALDAAEGLSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSK 850
Query: 242 T-HVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLV 300
T + + G+ GY APEY + ++ KSDIYSFGVVLLEL+TG++ D+ G++++
Sbjct: 851 TPEAMSGIAGSCGYIAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTDSEL--GDKDMA 908
Query: 301 SWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
W D+ ++DP L +F + + I I +C P RP + +V+ L+
Sbjct: 909 KWVCTAL-DKCGLEPVIDPKLDLKFKEE-ISKVIHIGLLCTSPLPLNRPSMRKVVIMLQ 965
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 191 bits (484), Expect = 4e-48
Identities = 110/287 (38%), Positives = 165/287 (57%), Gaps = 7/287 (2%)
Query: 77 ELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDG-RQGFQEFVMEVLMLSL 135
++ +AT N + +IG+G G +YK L+ + AVK+L G + G V E+ +
Sbjct: 808 KVLEATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGK 867
Query: 136 LHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARG 195
+ H NL++L + + L++Y YM GSL D L E + K PL+WSTR +AVG A G
Sbjct: 868 VRHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPK-PLDWSTRHNIAVGTAHG 926
Query: 196 LEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYC 255
L YLH DP +++RD+K NILLD++ P +SDFG+AKL + S V GT GY
Sbjct: 927 LAYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYM 986
Query: 256 APEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGH 315
APE A + + +SD+YS+GVVLLEL+T ++A+D S GE ++V W R ++
Sbjct: 987 APENAFTTVKSRESDVYSYGVVLLELITRKKALDPSFN-GETDIVGWVRSVWTQTGEIQK 1045
Query: 316 MVDP-LLQGRFPSRCLHQ---AIAITAMCLQEQPKFRPLITDIVVAL 358
+VDP LL S + Q A+++ C +++ RP + D+V L
Sbjct: 1046 IVDPSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
>IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (EC
2.7.1.37) (IRAK-4)
Length = 459
Score = 187 bits (476), Expect = 3e-47
Identities = 122/302 (40%), Positives = 167/302 (54%), Gaps = 19/302 (6%)
Query: 72 SFGFRELADATRNFKE------ANLIGEGGFGKVYKGRLTTGEAVAVKQLSH----DGRQ 121
SF F EL T NF E N +GEGGFG VYKG VAVK+L +
Sbjct: 167 SFSFHELKSITNNFDEQPASAGGNRMGEGGFGVVYKG-CVNNTIVAVKKLGAMVEISTEE 225
Query: 122 GFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLN 181
Q+F E+ +++ H NLV L+G+ +D D LVY YMP GSL D L L PL+
Sbjct: 226 LKQQFDQEIKVMATCQHENLVELLGFSSDSDNLCLVYAYMPNGSLLDRLSCLD-GTPPLS 284
Query: 182 WSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDN 241
W TR KVA G A G+ +LH + I+RD+KSANILLD +F K+SDFGLA+
Sbjct: 285 WHTRCKVAQGTANGIRFLH---ENHHIHRDIKSANILLDKDFTAKISDFGLARASARLAQ 341
Query: 242 THVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVS 301
T +++R++GT Y APE A+ G++T KSDIYSFGVVLLEL+TG A+D +R P Q L+
Sbjct: 342 TVMTSRIVGTTAYMAPE-ALRGEITPKSDIYSFGVVLLELITGLAAVDENREP--QLLLD 398
Query: 302 WARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYL 361
+ + D + P+ + + + CL E+ RP I + L+ +
Sbjct: 399 IKEEIEDEEKTIEDYTDEKMSDADPA-SVEAMYSAASQCLHEKKNRRPDIAKVQQLLQEM 457
Query: 362 AS 363
++
Sbjct: 458 SA 459
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,303,671
Number of Sequences: 164201
Number of extensions: 2004831
Number of successful extensions: 10906
Number of sequences better than 10.0: 1655
Number of HSP's better than 10.0 without gapping: 1095
Number of HSP's successfully gapped in prelim test: 560
Number of HSP's that attempted gapping in prelim test: 6687
Number of HSP's gapped (non-prelim): 2101
length of query: 385
length of database: 59,974,054
effective HSP length: 112
effective length of query: 273
effective length of database: 41,583,542
effective search space: 11352306966
effective search space used: 11352306966
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0193.1


