

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0191.7
(680 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
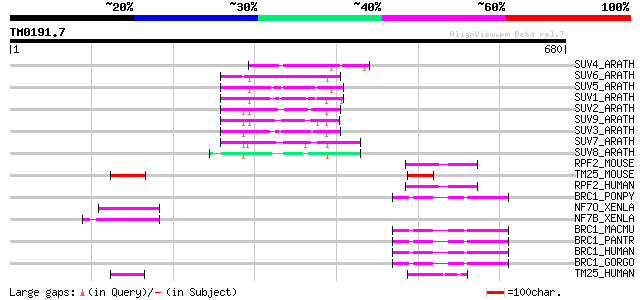
Score E
Sequences producing significant alignments: (bits) Value
SUV4_ARATH (Q8GZB6) Histone-lysine N-methyltransferase, H3 lysin... 83 2e-15
SUV6_ARATH (Q8VZ17) Histone-lysine N-methyltransferase, H3 lysin... 79 4e-14
SUV5_ARATH (O82175) Histone-lysine N-methyltransferase, H3 lysin... 74 2e-12
SUV1_ARATH (Q9FF80) Histone-lysine N-methyltransferase, H3 lysin... 71 1e-11
SUV2_ARATH (O22781) Probable histone-lysine N-methyltransferase,... 69 5e-11
SUV9_ARATH (Q9T0G7) Probable histone-lysine N-methyltransferase,... 67 2e-10
SUV3_ARATH (Q9C5P4) Histone-lysine N-methyltransferase, H3 lysin... 64 1e-09
SUV7_ARATH (Q9C5P1) Histone-lysine N-methyltransferase, H3 lysin... 61 1e-08
SUV8_ARATH (Q9C5P0) Histone-lysine N-methyltransferase, H3 lysin... 55 8e-07
RPF2_MOUSE (Q9CYB0) Ret finger protein 2 (Putative tumor suppres... 55 8e-07
TM25_MOUSE (Q61510) Tripartite motif-containing protein 25 (Zinc... 54 1e-06
RPF2_HUMAN (O60858) Ret finger protein 2 (Leukemia associated pr... 54 1e-06
BRC1_PONPY (Q6J6J0) Breast cancer type 1 susceptibility protein ... 52 4e-06
NF7O_XENLA (Q91431) Nuclear factor 7, ovary (xnf7-O) 52 5e-06
NF7B_XENLA (Q92021) Nuclear factor 7, brain (xnf7) (xnf7-B) 52 5e-06
BRC1_MACMU (Q6J6I9) Breast cancer type 1 susceptibility protein ... 50 1e-05
BRC1_PANTR (Q9GKK8) Breast cancer type 1 susceptibility protein ... 50 2e-05
BRC1_HUMAN (P38398) Breast cancer type 1 susceptibility protein 50 2e-05
BRC1_GORGO (Q6J6I8) Breast cancer type 1 susceptibility protein ... 50 2e-05
TM25_HUMAN (Q14258) Tripartite motif-containing protein 25 (Zinc... 49 4e-05
>SUV4_ARATH (Q8GZB6) Histone-lysine N-methyltransferase, H3 lysine-9
specific 4 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 4) (H3-K9-HMTase 4) (Suppressor of
variegation 3-9 homolog 4) (Su(var)3-9 homolog 4)
(KRYPTONITE protein)
Length = 624
Score = 83.2 bits (204), Expect = 2e-15
Identities = 62/157 (39%), Positives = 83/157 (52%), Gaps = 16/157 (10%)
Query: 293 AQSVALSGGYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKG 352
A S+ +SG Y DD D+ + YTG GG +L+GNKR K DQ E N AL+ C
Sbjct: 198 AVSIVMSGQYEDDLDNADTVTYTGQGGHNLTGNKRQIK----DQLLERGNLALKHCCEYN 253
Query: 353 YPVRVVRSHKEKRSSYAPESGVRYDGVYRIEKCWRKNGIQ---VCRYLFVRCDNEPAPWT 409
PVRV R H K SSY + YDG+Y++EK W + G+ V +Y R + +P T
Sbjct: 254 VPVRVTRGHNCK-SSYT-KRVYTYDGLYKVEKFWAQKGVSGFTVYKYRLKRLEGQP-ELT 310
Query: 410 TDDVGDRPRPLP-KIDELKGAV--DIT---ERKGDPS 440
TD V +P E++G V DI+ E KG P+
Sbjct: 311 TDQVNFVAGRIPTSTSEIEGLVCEDISGGLEFKGIPA 347
>SUV6_ARATH (Q8VZ17) Histone-lysine N-methyltransferase, H3 lysine-9
specific 6 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 6) (H3-K9-HMTase 6) (Suppressor of
variegation 3-9 homolog 6) (Su(var)3-9 homolog 6)
Length = 790
Score = 79.0 bits (193), Expect = 4e-14
Identities = 54/155 (34%), Positives = 76/155 (48%), Gaps = 9/155 (5%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYG----AQSVALSGGYIDDEDHGEWFLY 314
GV VG+ ++ RME G H P AGI YG A S+ SGGY D D+ + Y
Sbjct: 334 GVEVGDEFQYRMELNILGIHKPSQAGI-DYMKYGKAKVATSIVASGGYDDHLDNSDVLTY 392
Query: 315 TGSGGRDLSGNKRTNKLQS-FDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPESG 373
TG GG + K+ +L+ DQK N AL S K PVRV+R + +
Sbjct: 393 TGQGGNVMQVKKKGEELKEPEDQKLITGNLALATSIEKQTPVRVIRGKHKSTHDKSKGGN 452
Query: 374 VRYDGVYRIEKCWRK---NGIQVCRYLFVRCDNEP 405
YDG+Y +EK W++ +G+ V ++ R +P
Sbjct: 453 YVYDGLYLVEKYWQQVGSHGMNVFKFQLRRIPGQP 487
>SUV5_ARATH (O82175) Histone-lysine N-methyltransferase, H3 lysine-9
specific 5 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 5) (H3-K9-HMTase 5) (Suppressor of
variegation 3-9 homolog 5) (Su(var)3-9 homolog 5)
Length = 794
Score = 73.6 bits (179), Expect = 2e-12
Identities = 57/160 (35%), Positives = 80/160 (49%), Gaps = 14/160 (8%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYG----AQSVALSGGYIDDEDHGEWFLY 314
GV VG+ ++ RME G H P +GI G A S+ SGGY D D+ + +Y
Sbjct: 369 GVEVGDEFQYRMELNLLGIHRPSQSGIDYMKDDGGELVATSIVSSGGYNDVLDNSDVLIY 428
Query: 315 TGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEK--RSSYAPES 372
TG GG G K+ N+ DQ+ N AL+ S K PVRV+R K +SS ++
Sbjct: 429 TGQGGN--VGKKKNNEPPK-DQQLVTGNLALKNSINKKNPVRVIRGIKNTTLQSSVVAKN 485
Query: 373 GVRYDGVYRIEKCWRKNGIQ---VCRYLFVRCDNEP-APW 408
V YDG+Y +E+ W + G V ++ R +P PW
Sbjct: 486 YV-YDGLYLVEEYWEETGSHGKLVFKFKLRRIPGQPELPW 524
>SUV1_ARATH (Q9FF80) Histone-lysine N-methyltransferase, H3 lysine-9
specific 1 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of
variegation 3-9 homolog 1) (Su(var)3-9 homolog 1)
Length = 670
Score = 70.9 bits (172), Expect = 1e-11
Identities = 59/164 (35%), Positives = 81/164 (48%), Gaps = 22/164 (13%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGIAGQSTYG-------AQSVALSGGYIDDEDHGEW 311
GV +G+ + R E G H P +AGI G A S+ SG Y +DE + +
Sbjct: 215 GVEIGDVFFFRFEMCLVGLHSPSMAGIDYLVVKGETEEEPIATSIVSSGYYDNDEGNPDV 274
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPE 371
+YTG GG N +K QS DQK E N AL S R+ VRV+R KE +S+ +
Sbjct: 275 LIYTGQGG-----NADKDK-QSSDQKLERGNLALEKSLRRDSAVRVIRGLKE--ASHNAK 326
Query: 372 SGVRYDGVYRIEKCW---RKNGIQVCRYLFVRCDNEP---APWT 409
+ YDG+Y I++ W K+G +Y VR +P A WT
Sbjct: 327 IYI-YDGLYEIKESWVEKGKSGHNTFKYKLVRAPGQPPAFASWT 369
>SUV2_ARATH (O22781) Probable histone-lysine N-methyltransferase, H3
lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of
variegation 3-9 homolog 2) (Su(var)3-9 homolog 2)
Length = 651
Score = 68.6 bits (166), Expect = 5e-11
Identities = 55/157 (35%), Positives = 75/157 (47%), Gaps = 18/157 (11%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGI----AGQSTYG---AQSVALSGGYIDDEDHGEW 311
GV VG+ + RME G H AGI A +S G A S+ +SGGY DDED G+
Sbjct: 211 GVEVGDIFFYRMELCVLGLHGQTQAGIDCLTAERSATGEPIATSIVVSGGYEDDEDTGDV 270
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPE 371
+YTG GG+D + N Q+ N + S G VRV+R K + S
Sbjct: 271 LVYTGHGGQDHQHKQCDN------QRLVGGNLGMERSMHYGIEVRVIRGIKYENS--ISS 322
Query: 372 SGVRYDGVYRIEKCW---RKNGIQVCRYLFVRCDNEP 405
YDG+Y+I W K+G V ++ VR + +P
Sbjct: 323 KVYVYDGLYKIVDWWFAVGKSGFGVFKFRLVRIEGQP 359
>SUV9_ARATH (Q9T0G7) Probable histone-lysine N-methyltransferase, H3
lysine-9 specific 9 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 9) (H3-K9-HMTase 9) (Suppressor of
variegation 3-9 homolog 9) (Su(var)3-9 homolog 9)
Length = 650
Score = 66.6 bits (161), Expect = 2e-10
Identities = 56/159 (35%), Positives = 75/159 (46%), Gaps = 24/159 (15%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGI----AGQSTYG---AQSVALSGGYIDDEDHGEW 311
GV VG+ + R E G H +GI S+ G A SV +SGGY DD+D G+
Sbjct: 209 GVQVGDIFFFRFELCVMGLHGHPQSGIDFLTGSLSSNGEPIATSVIVSGGYEDDDDQGDV 268
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPE 371
+YTG GG+D G Q+ Q+ E N A+ S G VRV+R K Y E
Sbjct: 269 IMYTGQGGQDRLGR------QAEHQRLEGGNLAMERSMYYGIEVRVIRGLK-----YENE 317
Query: 372 SGVR---YDGVYRIEKCW---RKNGIQVCRYLFVRCDNE 404
R YDG++RI W K+G V +Y R + +
Sbjct: 318 VSSRVYVYDGLFRIVDSWFDVGKSGFGVFKYRLERIEGQ 356
>SUV3_ARATH (Q9C5P4) Histone-lysine N-methyltransferase, H3 lysine-9
specific 3 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 3) (H3-K9-HMTase 3) (Suppressor of
variegation 3-9 homolog 3) (Su(var)3-9 homolog 3)
Length = 669
Score = 64.3 bits (155), Expect = 1e-09
Identities = 54/157 (34%), Positives = 73/157 (46%), Gaps = 18/157 (11%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGI------AGQSTYG-AQSVALSGGYIDDEDHGEW 311
G+ VG+ + R+E G H+ +AGI AG A S+ SG Y + E
Sbjct: 212 GIEVGDIFFSRIEMCLVGLHMQTMAGIDYIISKAGSDEESLATSIVSSGRYEGEAQDPES 271
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRSHKEKRSSYAPE 371
+Y+G GG N N+ Q+ DQK E N AL S RKG VRVVR ++ S
Sbjct: 272 LIYSGQGG-----NADKNR-QASDQKLERGNLALENSLRKGNGVRVVRGEEDAASKTG-- 323
Query: 372 SGVRYDGVYRIEKCW---RKNGIQVCRYLFVRCDNEP 405
YDG+Y I + W K+G +Y VR +P
Sbjct: 324 KIYIYDGLYSISESWVEKGKSGCNTFKYKLVRQPGQP 360
>SUV7_ARATH (Q9C5P1) Histone-lysine N-methyltransferase, H3 lysine-9
specific 7 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 7) (H3-K9-HMTase 7) (Suppressor of
variegation 3-9 homolog 7) (Su(var)3-9 homolog 7)
Length = 693
Score = 60.8 bits (146), Expect = 1e-08
Identities = 54/186 (29%), Positives = 81/186 (43%), Gaps = 27/186 (14%)
Query: 259 GVLVGETWEDRMECRQWGAHLPHVAGI----AGQST---YGAQSVALSGGYIDDEDHGEW 311
G+ VG+ + E G H + GI A +S + A V +G Y + + +
Sbjct: 231 GIHVGDIFYYWGEMCLVGLHKSNYGGIDFFTAAESAVEGHAAMCVVTAGQYDGETEGLDT 290
Query: 312 FLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVVRS----HKEKRSS 367
+Y+G GG D+ GN R DQ+ + N AL S KG VRVVR H+ +
Sbjct: 291 LIYSGQGGTDVYGNAR-------DQEMKGGNLALEASVSKGNDVRVVRGVIHPHENNQKI 343
Query: 368 YAPESGVRYDGVYRIEKCWR---KNGIQVCRYLFVRCDNEPAPWTTDDVGDRPRPLPKID 424
Y YDG+Y + K W K+G + R+ VR N+P + + R ID
Sbjct: 344 YI------YDGMYLVSKFWTVTGKSGFKEFRFKLVRKPNQPPAYAIWKTVENLRNHDLID 397
Query: 425 ELKGAV 430
+G +
Sbjct: 398 SRQGFI 403
>SUV8_ARATH (Q9C5P0) Histone-lysine N-methyltransferase, H3 lysine-9
specific 8 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 8) (H3-K9-HMTase 8) (Suppressor of
variegation 3-9 homolog 8) (Su(var)3-9 homolog 8)
Length = 755
Score = 54.7 bits (130), Expect = 8e-07
Identities = 53/195 (27%), Positives = 78/195 (39%), Gaps = 36/195 (18%)
Query: 246 GPIPAENDPTRNQGVLVGETWEDRMECRQWGAHLPHVAGI-------AGQSTYGAQSVAL 298
GPIP GV VG+ + E G H GI +G A SV
Sbjct: 310 GPIP---------GVQVGDIFYYWCEMCLVGLHRNTAGGIDSLLAKESGVDGPAATSVVT 360
Query: 299 SGGYIDDEDHGEWFLYTGSGGRDLSGNKRTNKLQSFDQKFENMNEALRLSCRKGYPVRVV 358
SG Y ++ + E +Y+G GG+ DQ + N AL S R+ VRV+
Sbjct: 361 SGKYDNETEDLETLIYSGHGGKPC------------DQVLQRGNRALEASVRRRNEVRVI 408
Query: 359 RSHKEKRSSYAPESGVRYDGVYRIEKCWR---KNGIQVCRYLFVRCDNEPAPWTTDDVGD 415
R Y E YDG+Y + CW+ K+G + R+ +R +P + + +
Sbjct: 409 RG-----ELYNNEKVYIYDGLYLVSDCWQVTGKSGFKEYRFKLLRKPGQPPGYAIWKLVE 463
Query: 416 RPRPLPKIDELKGAV 430
R ID +G +
Sbjct: 464 NLRNHELIDPRQGFI 478
>RPF2_MOUSE (Q9CYB0) Ret finger protein 2 (Putative tumor suppressor
RFP2) (Tripartite motif-containing protein 13)
Length = 407
Score = 54.7 bits (130), Expect = 8e-07
Identities = 30/89 (33%), Positives = 43/89 (47%), Gaps = 11/89 (12%)
Query: 486 EKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNV 545
E L ++ C IC + P PC+HNFCK CLEG G R+ + +
Sbjct: 2 ELLEEDLTCPICCSLFDDPRVLPCSHNFCKKCLEGLLEGNV----------RNSLWRPSP 51
Query: 546 MKCPSCSNDI-ADFLQNPQVNREMMGVVE 573
KCP+C + A + + QVN + G+VE
Sbjct: 52 FKCPTCRKETSATGVNSLQVNYSLKGIVE 80
Score = 44.3 bits (103), Expect = 0.001
Identities = 21/75 (28%), Positives = 32/75 (42%), Gaps = 9/75 (12%)
Query: 117 LDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQGKRT---------CANCRRD 167
+++ + + C C L D P PC HNFC KC E + R C CR++
Sbjct: 1 MELLEEDLTCPICCSLFDDPRVLPCSHNFCKKCLEGLLEGNVRNSLWRPSPFKCPTCRKE 60
Query: 168 IPAKIASQPRINSQL 182
A + ++N L
Sbjct: 61 TSATGVNSLQVNYSL 75
>TM25_MOUSE (Q61510) Tripartite motif-containing protein 25 (Zinc
finger protein 147) (Estrogen responsive finger protein)
(Efp)
Length = 634
Score = 54.3 bits (129), Expect = 1e-06
Identities = 23/45 (51%), Positives = 29/45 (64%), Gaps = 2/45 (4%)
Query: 124 INCSFCMQLPDRPVTTPCGHNFCLKCF-EKWIGQG-KRTCANCRR 166
++CS C++L PVTTPCGHNFC C E W+ QG C CR+
Sbjct: 11 LSCSVCLELFKEPVTTPCGHNFCTSCLDETWVVQGPPYRCPQCRK 55
Score = 45.1 bits (105), Expect = 6e-04
Identities = 15/32 (46%), Positives = 22/32 (67%)
Query: 488 LLKEFGCNICHKVLSSPLTTPCAHNFCKVCLE 519
L +E C++C ++ P+TTPC HNFC CL+
Sbjct: 7 LAEELSCSVCLELFKEPVTTPCGHNFCTSCLD 38
>RPF2_HUMAN (O60858) Ret finger protein 2 (Leukemia associated
protein 5) (B-cell chronic lymphocytic leukemia tumor
suppressor Leu5) (Putative tumor suppressor RFP2)
(Tripartite motif-containing protein 13)
Length = 407
Score = 53.9 bits (128), Expect = 1e-06
Identities = 30/89 (33%), Positives = 42/89 (46%), Gaps = 11/89 (12%)
Query: 486 EKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNV 545
E L ++ C IC + P PC+HNFCK CLEG G R+ +
Sbjct: 2 ELLEEDLTCPICCSLFDDPRVLPCSHNFCKKCLEGILEGSV----------RNSLWRPAP 51
Query: 546 MKCPSCSNDI-ADFLQNPQVNREMMGVVE 573
KCP+C + A + + QVN + G+VE
Sbjct: 52 FKCPTCRKETSATGINSLQVNYSLKGIVE 80
Score = 43.9 bits (102), Expect = 0.001
Identities = 27/97 (27%), Positives = 40/97 (40%), Gaps = 15/97 (15%)
Query: 117 LDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQGKRT---------CANCRRD 167
+++ + + C C L D P PC HNFC KC E + R C CR++
Sbjct: 1 MELLEEDLTCPICCSLFDDPRVLPCSHNFCKKCLEGILEGSVRNSLWRPAPFKCPTCRKE 60
Query: 168 IPAKIASQPRINSQLAIAIRLAKLAKSEGKVSGEPKI 204
S INS L + L + + K+ PK+
Sbjct: 61 -----TSATGINS-LQVNYSLKGIVEKYNKIKISPKM 91
>BRC1_PONPY (Q6J6J0) Breast cancer type 1 susceptibility protein
homolog
Length = 1863
Score = 52.4 bits (124), Expect = 4e-06
Identities = 40/145 (27%), Positives = 68/145 (46%), Gaps = 25/145 (17%)
Query: 470 DGTRVRVKKTKNV-SLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFI 528
D + VRV++ +NV + +K+L+ C IC +++ P++T C H FCK C+
Sbjct: 2 DLSAVRVEEVQNVINAMQKILE---CPICLELIKEPVSTKCDHIFCKFCML--------- 49
Query: 529 RKRACEGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQIA--QDENSE 586
+ L +K +CP C NDI ++ Q + +VE L I Q +
Sbjct: 50 --------KLLNQKKGPSQCPLCKNDITK--RSLQESTRFSQLVEELLKIICAFQLDTGL 99
Query: 587 QVENSEELSDKSDEIVKNVPDEVEV 611
Q NS + K + +++ DEV +
Sbjct: 100 QYANSYNFAKKENNSPEHLKDEVSI 124
Score = 42.7 bits (99), Expect = 0.003
Identities = 21/69 (30%), Positives = 32/69 (45%), Gaps = 2/69 (2%)
Query: 116 ILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQ--GKRTCANCRRDIPAKIA 173
+++ + C C++L PV+T C H FC C K + Q G C C+ DI +
Sbjct: 14 VINAMQKILECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSL 73
Query: 174 SQPRINSQL 182
+ SQL
Sbjct: 74 QESTRFSQL 82
>NF7O_XENLA (Q91431) Nuclear factor 7, ovary (xnf7-O)
Length = 610
Score = 52.0 bits (123), Expect = 5e-06
Identities = 26/76 (34%), Positives = 36/76 (47%), Gaps = 1/76 (1%)
Query: 109 KPPAAGGILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEK-WIGQGKRTCANCRRD 167
K A+ G F + C C++L PV CGHNFC C +K W GQ C C+
Sbjct: 129 KTAASLGAAGDFAEELTCPLCVELFKDPVMVACGHNFCRSCIDKVWEGQSSFACPECKES 188
Query: 168 IPAKIASQPRINSQLA 183
I + + R+ + LA
Sbjct: 189 ITDRKYTINRVLANLA 204
Score = 42.4 bits (98), Expect = 0.004
Identities = 24/105 (22%), Positives = 41/105 (38%), Gaps = 24/105 (22%)
Query: 453 KKPPPASKKSIDVINPKDGTRVRVKKTKNVSLKEKLLKEFGCNICHKVLSSPLTTPCAHN 512
++P P KK+ + KD + K ++ +E C +C ++ P+ C HN
Sbjct: 111 EEPEPEPKKA--KVEDKDAS----KTAASLGAAGDFAEELTCPLCVELFKDPVMVACGHN 164
Query: 513 FCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNVMKCPSCSNDIAD 557
FC+ C++ + G+S CP C I D
Sbjct: 165 FCRSCIDKVWEGQS------------------SFACPECKESITD 191
>NF7B_XENLA (Q92021) Nuclear factor 7, brain (xnf7) (xnf7-B)
Length = 609
Score = 52.0 bits (123), Expect = 5e-06
Identities = 29/95 (30%), Positives = 44/95 (45%), Gaps = 7/95 (7%)
Query: 90 DQQKAKKKQELVGGPSSGEKPPAAGGILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKC 149
+ +KAK +++ K ++ G F + C C++L PV CGHNFC C
Sbjct: 115 EPKKAKVEEK------DASKNASSLGAAGDFAEELTCPLCVELFKDPVMVACGHNFCRSC 168
Query: 150 FEK-WIGQGKRTCANCRRDIPAKIASQPRINSQLA 183
+K W GQ C CR I + + R+ + LA
Sbjct: 169 IDKAWEGQSSFACPECRESITDRKYTINRVLANLA 203
Score = 44.3 bits (103), Expect = 0.001
Identities = 25/105 (23%), Positives = 42/105 (39%), Gaps = 24/105 (22%)
Query: 453 KKPPPASKKSIDVINPKDGTRVRVKKTKNVSLKEKLLKEFGCNICHKVLSSPLTTPCAHN 512
++P P KK+ + KD + K ++ +E C +C ++ P+ C HN
Sbjct: 110 EEPEPEPKKA--KVEEKDAS----KNASSLGAAGDFAEELTCPLCVELFKDPVMVACGHN 163
Query: 513 FCKVCLEGAFSGKSFIRKRACEGGRSLRAQKNVMKCPSCSNDIAD 557
FC+ C++ A+ G+S CP C I D
Sbjct: 164 FCRSCIDKAWEGQS------------------SFACPECRESITD 190
>BRC1_MACMU (Q6J6I9) Breast cancer type 1 susceptibility protein
homolog
Length = 1863
Score = 50.4 bits (119), Expect = 1e-05
Identities = 39/145 (26%), Positives = 68/145 (46%), Gaps = 25/145 (17%)
Query: 470 DGTRVRVKKTKNV-SLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFI 528
D + VRV++ +NV + +K+L+ C IC +++ P++T C H FC+ C+
Sbjct: 2 DLSAVRVEEVQNVINAMQKILE---CPICLELIKEPVSTKCDHIFCRFCML--------- 49
Query: 529 RKRACEGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQI--AQDENSE 586
+ L +K +CP C NDI ++ Q + +VE L I Q +
Sbjct: 50 --------KLLNQKKGPSQCPLCKNDITK--RSLQESTRFSQLVEELLKIIHAFQLDTGL 99
Query: 587 QVENSEELSDKSDEIVKNVPDEVEV 611
Q NS + K + +++ DEV +
Sbjct: 100 QFANSYNFAKKENHSPEHLKDEVSI 124
Score = 42.7 bits (99), Expect = 0.003
Identities = 21/69 (30%), Positives = 32/69 (45%), Gaps = 2/69 (2%)
Query: 116 ILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQ--GKRTCANCRRDIPAKIA 173
+++ + C C++L PV+T C H FC C K + Q G C C+ DI +
Sbjct: 14 VINAMQKILECPICLELIKEPVSTKCDHIFCRFCMLKLLNQKKGPSQCPLCKNDITKRSL 73
Query: 174 SQPRINSQL 182
+ SQL
Sbjct: 74 QESTRFSQL 82
>BRC1_PANTR (Q9GKK8) Breast cancer type 1 susceptibility protein
homolog
Length = 1863
Score = 50.1 bits (118), Expect = 2e-05
Identities = 38/145 (26%), Positives = 68/145 (46%), Gaps = 25/145 (17%)
Query: 470 DGTRVRVKKTKNV-SLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFI 528
D + +RV++ +NV + +K+L+ C IC +++ P++T C H FCK C+
Sbjct: 2 DLSALRVEEVQNVINAMQKILE---CPICLELIKEPVSTKCDHIFCKFCML--------- 49
Query: 529 RKRACEGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQIA--QDENSE 586
+ L +K +CP C NDI ++ Q + +VE L I Q +
Sbjct: 50 --------KLLNQKKGPSQCPLCKNDITK--RSLQESTRFSQLVEELLKIICAFQLDTGL 99
Query: 587 QVENSEELSDKSDEIVKNVPDEVEV 611
+ NS + K + +++ DEV +
Sbjct: 100 EYANSYNFAKKENNSPEHLKDEVSI 124
Score = 42.7 bits (99), Expect = 0.003
Identities = 21/69 (30%), Positives = 32/69 (45%), Gaps = 2/69 (2%)
Query: 116 ILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQ--GKRTCANCRRDIPAKIA 173
+++ + C C++L PV+T C H FC C K + Q G C C+ DI +
Sbjct: 14 VINAMQKILECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSL 73
Query: 174 SQPRINSQL 182
+ SQL
Sbjct: 74 QESTRFSQL 82
>BRC1_HUMAN (P38398) Breast cancer type 1 susceptibility protein
Length = 1863
Score = 50.1 bits (118), Expect = 2e-05
Identities = 38/145 (26%), Positives = 68/145 (46%), Gaps = 25/145 (17%)
Query: 470 DGTRVRVKKTKNV-SLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFI 528
D + +RV++ +NV + +K+L+ C IC +++ P++T C H FCK C+
Sbjct: 2 DLSALRVEEVQNVINAMQKILE---CPICLELIKEPVSTKCDHIFCKFCML--------- 49
Query: 529 RKRACEGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQIA--QDENSE 586
+ L +K +CP C NDI ++ Q + +VE L I Q +
Sbjct: 50 --------KLLNQKKGPSQCPLCKNDITK--RSLQESTRFSQLVEELLKIICAFQLDTGL 99
Query: 587 QVENSEELSDKSDEIVKNVPDEVEV 611
+ NS + K + +++ DEV +
Sbjct: 100 EYANSYNFAKKENNSPEHLKDEVSI 124
Score = 42.7 bits (99), Expect = 0.003
Identities = 21/69 (30%), Positives = 32/69 (45%), Gaps = 2/69 (2%)
Query: 116 ILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQ--GKRTCANCRRDIPAKIA 173
+++ + C C++L PV+T C H FC C K + Q G C C+ DI +
Sbjct: 14 VINAMQKILECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSL 73
Query: 174 SQPRINSQL 182
+ SQL
Sbjct: 74 QESTRFSQL 82
>BRC1_GORGO (Q6J6I8) Breast cancer type 1 susceptibility protein
homolog
Length = 1863
Score = 50.1 bits (118), Expect = 2e-05
Identities = 38/145 (26%), Positives = 68/145 (46%), Gaps = 25/145 (17%)
Query: 470 DGTRVRVKKTKNV-SLKEKLLKEFGCNICHKVLSSPLTTPCAHNFCKVCLEGAFSGKSFI 528
D + +RV++ +NV + +K+L+ C IC +++ P++T C H FCK C+
Sbjct: 2 DLSALRVEEVQNVINAMQKILE---CPICLELIKEPVSTKCDHIFCKFCML--------- 49
Query: 529 RKRACEGGRSLRAQKNVMKCPSCSNDIADFLQNPQVNREMMGVVESLQSQIA--QDENSE 586
+ L +K +CP C NDI ++ Q + +VE L I Q +
Sbjct: 50 --------KLLNQKKGPSQCPLCKNDITK--RSLQESTRFSQLVEELLKIICAFQLDTGL 99
Query: 587 QVENSEELSDKSDEIVKNVPDEVEV 611
+ NS + K + +++ DEV +
Sbjct: 100 EYANSYNFAKKENNSPEHLKDEVSI 124
Score = 42.7 bits (99), Expect = 0.003
Identities = 21/69 (30%), Positives = 32/69 (45%), Gaps = 2/69 (2%)
Query: 116 ILDIFDGSINCSFCMQLPDRPVTTPCGHNFCLKCFEKWIGQ--GKRTCANCRRDIPAKIA 173
+++ + C C++L PV+T C H FC C K + Q G C C+ DI +
Sbjct: 14 VINAMQKILECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSL 73
Query: 174 SQPRINSQL 182
+ SQL
Sbjct: 74 QESTRFSQL 82
>TM25_HUMAN (Q14258) Tripartite motif-containing protein 25 (Zinc
finger protein 147) (Estrogen responsive finger protein)
(Efp)
Length = 630
Score = 48.9 bits (115), Expect = 4e-05
Identities = 22/44 (50%), Positives = 26/44 (59%), Gaps = 2/44 (4%)
Query: 124 INCSFCMQLPDRPVTTPCGHNFCLKCF-EKWIGQGK-RTCANCR 165
++CS C++ PVTTPCGHNFC C E W QG C CR
Sbjct: 11 LSCSICLEPFKEPVTTPCGHNFCGSCLNETWAVQGSPYLCPQCR 54
Score = 45.4 bits (106), Expect = 5e-04
Identities = 28/77 (36%), Positives = 37/77 (47%), Gaps = 9/77 (11%)
Query: 488 LLKEFGCNICHKVLSSPLTTPCAHNFCKVCLE--GAFSGKSFIRKRACEGGRSLRAQ--K 543
L +E C+IC + P+TTPC HNFC CL A G ++ + C R Q K
Sbjct: 7 LAEELSCSICLEPFKEPVTTPCGHNFCGSCLNETWAVQGSPYLCPQ-CRAVYQARPQLHK 65
Query: 544 NVMKCPSCSNDIADFLQ 560
N + C N + FLQ
Sbjct: 66 NTVLC----NVVEQFLQ 78
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.134 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 89,826,736
Number of Sequences: 164201
Number of extensions: 4259839
Number of successful extensions: 13541
Number of sequences better than 10.0: 318
Number of HSP's better than 10.0 without gapping: 158
Number of HSP's successfully gapped in prelim test: 162
Number of HSP's that attempted gapping in prelim test: 12905
Number of HSP's gapped (non-prelim): 685
length of query: 680
length of database: 59,974,054
effective HSP length: 117
effective length of query: 563
effective length of database: 40,762,537
effective search space: 22949308331
effective search space used: 22949308331
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0191.7


