

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
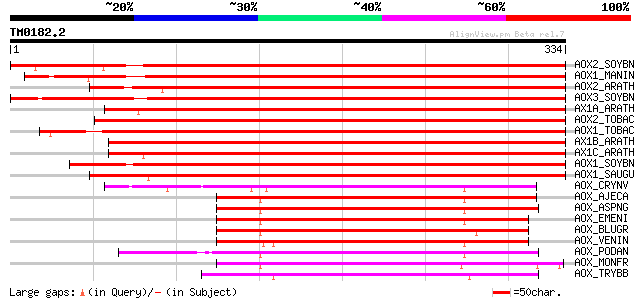
Query= TM0182.2
(334 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AOX2_SOYBN (Q41266) Alternative oxidase 2, mitochondrial precurs... 531 e-151
AOX1_MANIN (Q40294) Alternative oxidase, mitochondrial precursor... 495 e-140
AOX2_ARATH (O22049) Alternative oxidase 2, mitochondrial precurs... 457 e-128
AOX3_SOYBN (O03376) Alternative oxidase 3, mitochondrial precurs... 446 e-125
AX1A_ARATH (Q39219) Alternative oxidase 1a, mitochondrial precur... 442 e-124
AOX2_TOBAC (Q40578) Alternative oxidase 2, mitochondrial precurs... 431 e-120
AOX1_TOBAC (Q41224) Alternative oxidase 1, mitochondrial precurs... 430 e-120
AX1B_ARATH (O23913) Alternative oxidase 1b, mitochondrial precur... 427 e-119
AX1C_ARATH (O22048) Alternative oxidase 1c, mitochondrial precur... 424 e-118
AOX1_SOYBN (Q07185) Alternative oxidase 1, mitochondrial precurs... 422 e-118
AOX1_SAUGU (P22185) Alternative oxidase, mitochondrial precursor... 419 e-117
AOX_CRYNV (Q8NKE2) Alternative oxidase, mitochondrial precursor ... 211 3e-54
AOX_AJECA (Q9Y711) Alternative oxidase, mitochondrial precursor ... 203 6e-52
AOX_ASPNG (O74180) Alternative oxidase, mitochondrial precursor ... 199 1e-50
AOX_EMENI (Q9P959) Alternative oxidase, mitochondrial precursor ... 197 2e-50
AOX_BLUGR (Q8X1N9) Alternative oxidase, mitochondrial precursor ... 186 9e-47
AOX_VENIN (Q9P429) Alternative oxidase, mitochondrial precursor ... 185 1e-46
AOX_PODAN (Q9C206) Alternative oxidase, mitochondrial precursor ... 184 3e-46
AOX_MONFR (Q96UR9) Alternative oxidase, mitochondrial precursor ... 182 1e-45
AOX_TRYBB (Q26710) Alternative oxidase, mitochondrial precursor ... 181 3e-45
>AOX2_SOYBN (Q41266) Alternative oxidase 2, mitochondrial precursor
(EC 1.-.-.-)
Length = 333
Score = 531 bits (1368), Expect = e-151
Identities = 268/343 (78%), Positives = 286/343 (83%), Gaps = 19/343 (5%)
Query: 1 MKHLALSYALRRAL------NCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWNRMMSSQ 54
MK AL+ +RRAL N NR G A+ A E R G NG F W R M S
Sbjct: 1 MKLTALNSTVRRALLNGRNQNGNRLGSAALMPYAAAETRLLCAGGANGWFFYWKRTMVSP 60
Query: 55 A---APEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPW 111
A PE+EK++EKA+ E SVV SSYWGISRPK+ REDGTEWPWNCFMPW
Sbjct: 61 AEAKVPEKEKEKEKAKAEK----------SVVESSYWGISRPKVVREDGTEWPWNCFMPW 110
Query: 112 ETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGM 171
E+YR ++SIDLTKHHVPKN LDKVAYRTVKLLRIPTD+FF+RRYGCRAMMLETVAAVPGM
Sbjct: 111 ESYRSNVSIDLTKHHVPKNVLDKVAYRTVKLLRIPTDLFFKRRYGCRAMMLETVAAVPGM 170
Query: 172 VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFF 231
VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELV+PKWYER LVL VQGVFF
Sbjct: 171 VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVKPKWYERLLVLAVQGVFF 230
Query: 232 NAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDA 291
NAFFVLY+LSPKVAHR+VGYLEEEAIHSYTEYLKD+ESGAIENVPAPAIAIDYWRLPKDA
Sbjct: 231 NAFFVLYILSPKVAHRIVGYLEEEAIHSYTEYLKDLESGAIENVPAPAIAIDYWRLPKDA 290
Query: 292 TLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
LKDVITVIRADEAHHRDVNHFASDIHF GKELREAPAP+GYH
Sbjct: 291 RLKDVITVIRADEAHHRDVNHFASDIHFQGKELREAPAPIGYH 333
>AOX1_MANIN (Q40294) Alternative oxidase, mitochondrial precursor
(EC 1.-.-.-)
Length = 318
Score = 495 bits (1275), Expect = e-140
Identities = 242/328 (73%), Positives = 273/328 (82%), Gaps = 16/328 (4%)
Query: 10 LRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVFSC---WNRMMSSQAAPEEEKKEEKA 66
+R LN R+G + A +R V NG+ S W RM+S+ E + KE+K
Sbjct: 4 MRGLLNGGRYGNRYI--WTAISLRHPEVMEGNGLESAVMQWRRMLSNAGGAEAQVKEQKE 61
Query: 67 EKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTKHH 126
EK+ +VS+YWGISRPKITREDG+EWPWNCFMPWETYR DLSIDL KHH
Sbjct: 62 EKKD-----------AMVSNYWGISRPKITREDGSEWPWNCFMPWETYRSDLSIDLKKHH 110
Query: 127 VPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQ 186
VP+ F+DK AYRTVK+LR+PTD+FFQRRYGCRAMMLETVAAVPGMVGGMLLHL+SLRK +
Sbjct: 111 VPRTFMDKFAYRTVKILRVPTDIFFQRRYGCRAMMLETVAAVPGMVGGMLLHLKSLRKLE 170
Query: 187 QSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAH 246
QSGGWIKALLEEAENERMHLMTMVELVQPKWYER LVL VQGVFFN+FFVLY+LSPK+AH
Sbjct: 171 QSGGWIKALLEEAENERMHLMTMVELVQPKWYERLLVLAVQGVFFNSFFVLYVLSPKLAH 230
Query: 247 RVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITVIRADEAH 306
R+VGYLEEEAIHSYTEYLKDI+SGAI+N+PAPAIAIDYWRLPKDATLKDVITV+RADEAH
Sbjct: 231 RIVGYLEEEAIHSYTEYLKDIDSGAIKNIPAPAIAIDYWRLPKDATLKDVITVVRADEAH 290
Query: 307 HRDVNHFASDIHFHGKELREAPAPLGYH 334
HRDVNHFASD+ GKELR+APAP+GYH
Sbjct: 291 HRDVNHFASDVQVQGKELRDAPAPVGYH 318
>AOX2_ARATH (O22049) Alternative oxidase 2, mitochondrial precursor
(EC 1.-.-.-)
Length = 353
Score = 457 bits (1176), Expect = e-128
Identities = 219/288 (76%), Positives = 249/288 (86%), Gaps = 6/288 (2%)
Query: 49 RMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGI--SRPKITREDGTEWPWN 106
R M +A EKK+E + + + GSV V SYWGI ++ KITR+DG++WPWN
Sbjct: 70 RWMGMSSASAMEKKDENLTVK----KGQNGGGSVAVPSYWGIETAKMKITRKDGSDWPWN 125
Query: 107 CFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVA 166
CFMPWETY+ +LSIDL KHHVPKN DKVAYR VKLLRIPTD+FFQRRYGCRAMMLETVA
Sbjct: 126 CFMPWETYQANLSIDLKKHHVPKNIADKVAYRIVKLLRIPTDIFFQRRYGCRAMMLETVA 185
Query: 167 AVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTV 226
AVPGMVGGMLLHL+S+RKF+ SGGWIKALLEEAENERMHLMTM+ELV+PKWYER LV+ V
Sbjct: 186 AVPGMVGGMLLHLKSIRKFEHSGGWIKALLEEAENERMHLMTMMELVKPKWYERLLVMLV 245
Query: 227 QGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWR 286
QG+FFN+FFV Y++SP++AHRVVGYLEEEAIHSYTE+LKDI++G IENV APAIAIDYWR
Sbjct: 246 QGIFFNSFFVCYVISPRLAHRVVGYLEEEAIHSYTEFLKDIDNGKIENVAAPAIAIDYWR 305
Query: 287 LPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
LPKDATLKDV+TVIRADEAHHRDVNHFASDI GKELREA AP+GYH
Sbjct: 306 LPKDATLKDVVTVIRADEAHHRDVNHFASDIRNQGKELREAAAPIGYH 353
>AOX3_SOYBN (O03376) Alternative oxidase 3, mitochondrial precursor
(EC 1.-.-.-)
Length = 326
Score = 446 bits (1148), Expect = e-125
Identities = 219/335 (65%), Positives = 259/335 (76%), Gaps = 10/335 (2%)
Query: 1 MKHLALSYALRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWN-RMMSSQAAPEE 59
MK++ + A R L G + RQL + G F ++ R MS+ ++
Sbjct: 1 MKNVLVRSAARALLGGG--GRSYYRQLSTAAIVEQRHQHGGGAFGSFHLRRMSTLPEVKD 58
Query: 60 EKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLS 119
+ EEK + + + A VV+SYWGI+RPK+ REDGTEWPWNCFMPW++Y D+S
Sbjct: 59 QHSEEKKNEVNGTSNA-------VVTSYWGITRPKVRREDGTEWPWNCFMPWDSYHSDVS 111
Query: 120 IDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHL 179
ID+TKHH PK+ DKVA+R VK LR+ +D++F+ RYGC AMMLET+AAVPGMVGGMLLHL
Sbjct: 112 IDVTKHHTPKSLTDKVAFRAVKFLRVLSDIYFKERYGCHAMMLETIAAVPGMVGGMLLHL 171
Query: 180 RSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYL 239
+SLRKFQ SGGWIKALLEEAENERMHLMTMVELV+P W+ER L+ T QGVFFNAFFV YL
Sbjct: 172 KSLRKFQHSGGWIKALLEEAENERMHLMTMVELVKPSWHERLLIFTAQGVFFNAFFVFYL 231
Query: 240 LSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITV 299
LSPK AHR VGYLEEEA+ SYT++L IESG +ENVPAPAIAIDYWRLPKDATLKDV+TV
Sbjct: 232 LSPKAAHRFVGYLEEEAVISYTQHLNAIESGKVENVPAPAIAIDYWRLPKDATLKDVVTV 291
Query: 300 IRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
IRADEAHHRDVNHFASDIH GKEL+EAPAP+GYH
Sbjct: 292 IRADEAHHRDVNHFASDIHHQGKELKEAPAPIGYH 326
>AX1A_ARATH (Q39219) Alternative oxidase 1a, mitochondrial precursor
(EC 1.-.-.-)
Length = 354
Score = 442 bits (1138), Expect = e-124
Identities = 204/279 (73%), Positives = 241/279 (86%), Gaps = 2/279 (0%)
Query: 58 EEEKKEEKAEKESLRTEAK--KNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYR 115
EE+ ++K E ES +A N G ++SYWG+ KIT+EDG+EW WNCF PWETY+
Sbjct: 76 EEDANQKKTENESTGGDAAGGNNKGDKGIASYWGVEPNKITKEDGSEWKWNCFRPWETYK 135
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGM 175
D++IDL KHHVP FLD++AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGM
Sbjct: 136 ADITIDLKKHHVPTTFLDRIAYWTVKSLRWPTDLFFQRRYGCRAMMLETVAAVPGMVGGM 195
Query: 176 LLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFF 235
LLH +SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +PKWYER LV+TVQGVFFNA+F
Sbjct: 196 LLHCKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPKWYERALVITVQGVFFNAYF 255
Query: 236 VLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKD 295
+ YL+SPK AHR+VGYLEEEAIHSYTE+LK+++ G IENVPAPAIAIDYWRLP DATL+D
Sbjct: 256 LGYLISPKFAHRMVGYLEEEAIHSYTEFLKELDKGNIENVPAPAIAIDYWRLPADATLRD 315
Query: 296 VITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+ V+RADEAHHRDVNHFASDIH+ G+EL+EAPAP+GYH
Sbjct: 316 VVMVVRADEAHHRDVNHFASDIHYQGRELKEAPAPIGYH 354
>AOX2_TOBAC (Q40578) Alternative oxidase 2, mitochondrial precursor
(EC 1.-.-.-)
Length = 297
Score = 431 bits (1109), Expect = e-120
Identities = 199/284 (70%), Positives = 235/284 (82%), Gaps = 1/284 (0%)
Query: 52 SSQAAPEEEKKEEKAEKESLRTEAKKNDGSVV-VSSYWGISRPKITREDGTEWPWNCFMP 110
+S A +++ ++K E + G V SYWG+ K+T+EDGTEW WNCF P
Sbjct: 14 ASTVALNDKQHDKKVENGGAAASGGGDGGDEKSVVSYWGVPPSKVTKEDGTEWKWNCFRP 73
Query: 111 WETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPG 170
WETY+ DLSIDLTKHH P FLDK AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPG
Sbjct: 74 WETYKADLSIDLTKHHAPTTFLDKFAYWTVKALRYPTDIFFQRRYGCRAMMLETVAAVPG 133
Query: 171 MVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVF 230
MVGGMLLH +SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +P WYER LV VQGVF
Sbjct: 134 MVGGMLLHCKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPNWYERALVFAVQGVF 193
Query: 231 FNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKD 290
NA+FV YLLSPK+AHR+VGYLEEEAIHSYTE+LK+++ G IENVPAPAIAIDYWRLPKD
Sbjct: 194 INAYFVTYLLSPKLAHRIVGYLEEEAIHSYTEFLKELDKGNIENVPAPAIAIDYWRLPKD 253
Query: 291 ATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
+TL+DV+ V+RADEAHHRDVNHFA DIH+ G++L+++PAP+GYH
Sbjct: 254 STLRDVVLVVRADEAHHRDVNHFAPDIHYQGQQLKDSPAPIGYH 297
>AOX1_TOBAC (Q41224) Alternative oxidase 1, mitochondrial precursor
(EC 1.-.-.-)
Length = 353
Score = 430 bits (1106), Expect = e-120
Identities = 209/319 (65%), Positives = 246/319 (76%), Gaps = 12/319 (3%)
Query: 19 HGLTA--VRQLPATEVRRFLVSGENGVFSCWNRMMSSQAAPEEEKKEEKAEKESLRTEAK 76
HG+ A + T VR F V G S A +++ ++KAE S
Sbjct: 44 HGVPANPSEKAVVTWVRHFPVMGSRSAMSM---------ALNDKQHDKKAENGSAAATGG 94
Query: 77 KNDGSVV-VSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFLDKV 135
+ G V SYWG+ K+T+EDGTEW WNCF PWETY+ DLSIDLTKHH P FLDK
Sbjct: 95 GDGGDEKSVVSYWGVQPSKVTKEDGTEWKWNCFRPWETYKADLSIDLTKHHAPTTFLDKF 154
Query: 136 AYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKAL 195
AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGMLLH +SLR+F+QSGGWIK L
Sbjct: 155 AYWTVKSLRYPTDIFFQRRYGCRAMMLETVAAVPGMVGGMLLHCKSLRRFEQSGGWIKTL 214
Query: 196 LEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEE 255
L+EAENERMHLMT +E+ +P WYER LV VQGVFFNA+FV YLLSPK+AHR+VGYLEEE
Sbjct: 215 LDEAENERMHLMTFMEVAKPNWYERALVFAVQGVFFNAYFVTYLLSPKLAHRIVGYLEEE 274
Query: 256 AIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFAS 315
AIHSYTE+LK+++ G IENVPAPAIAIDY RLPKD+TL DV+ V+RADEAHHRDVNHFAS
Sbjct: 275 AIHSYTEFLKELDKGNIENVPAPAIAIDYCRLPKDSTLLDVVLVVRADEAHHRDVNHFAS 334
Query: 316 DIHFHGKELREAPAPLGYH 334
DIH+ G++L+++PAP+GYH
Sbjct: 335 DIHYQGQQLKDSPAPIGYH 353
>AX1B_ARATH (O23913) Alternative oxidase 1b, mitochondrial precursor
(EC 1.-.-.-)
Length = 325
Score = 427 bits (1097), Expect = e-119
Identities = 198/276 (71%), Positives = 236/276 (84%), Gaps = 1/276 (0%)
Query: 60 EKKEEKAEKESLRTEAKK-NDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDL 118
EKK+ EK S +A + N G ++ SYWG+ KIT+EDGTEW W+CF PWETY+ DL
Sbjct: 50 EKKKTTEEKGSSGGKADQGNKGEQLIVSYWGVKPMKITKEDGTEWKWSCFRPWETYKSDL 109
Query: 119 SIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLH 178
+IDL KHHVP DK+AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGML+H
Sbjct: 110 TIDLKKHHVPSTLPDKLAYWTVKSLRWPTDLFFQRRYGCRAMMLETVAAVPGMVGGMLVH 169
Query: 179 LRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLY 238
+SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +P WYER LV+ VQG+FFNA+F+ Y
Sbjct: 170 CKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPNWYERALVIAVQGIFFNAYFLGY 229
Query: 239 LLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVIT 298
L+SPK AHR+VGYLEEEAIHSYTE+LK++++G IENVPAPAIAIDYWRL DATL+DV+
Sbjct: 230 LISPKFAHRMVGYLEEEAIHSYTEFLKELDNGNIENVPAPAIAIDYWRLEADATLRDVVM 289
Query: 299 VIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+RADEAHHRDVNH+ASDIH+ G+EL+EAPAP+GYH
Sbjct: 290 VVRADEAHHRDVNHYASDIHYQGRELKEAPAPIGYH 325
>AX1C_ARATH (O22048) Alternative oxidase 1c, mitochondrial precursor
(EC 1.-.-.-)
Length = 329
Score = 424 bits (1089), Expect = e-118
Identities = 195/279 (69%), Positives = 234/279 (82%), Gaps = 4/279 (1%)
Query: 60 EKKEEKAEKESLRTEAKKND----GSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYR 115
EKK+ E+E K ND G ++ SYWG+ KIT+EDGTEW W+CF PWETY+
Sbjct: 51 EKKKTSEEEEGSGDGVKVNDQGNKGEQLIVSYWGVKPMKITKEDGTEWKWSCFRPWETYK 110
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGM 175
DL+IDL KHHVP DK+AY VK LR PTD+FFQRRYGCRA+MLETVAAVPGMVGGM
Sbjct: 111 ADLTIDLKKHHVPSTLPDKIAYWMVKSLRWPTDLFFQRRYGCRAIMLETVAAVPGMVGGM 170
Query: 176 LLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFF 235
L+H +SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +PKWYER LV++VQGVFFNA+
Sbjct: 171 LMHFKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPKWYERALVISVQGVFFNAYL 230
Query: 236 VLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKD 295
+ Y++SPK AHR+VGYLEEEAIHSYTE+LK++++G IENVPAPAIA+DYWRL DATL+D
Sbjct: 231 IGYIISPKFAHRMVGYLEEEAIHSYTEFLKELDNGNIENVPAPAIAVDYWRLEADATLRD 290
Query: 296 VITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+ V+RADEAHHRDVNH+ASDIH+ G EL+EAPAP+GYH
Sbjct: 291 VVMVVRADEAHHRDVNHYASDIHYQGHELKEAPAPIGYH 329
>AOX1_SOYBN (Q07185) Alternative oxidase 1, mitochondrial precursor
(EC 1.-.-.-)
Length = 321
Score = 422 bits (1085), Expect = e-118
Identities = 204/298 (68%), Positives = 238/298 (79%), Gaps = 4/298 (1%)
Query: 37 VSGENGVFSCWNRMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKIT 96
V G ++ R S+ A E+EK E+K S A N V+ SYWGI KIT
Sbjct: 28 VGGLRALYGGGVRSESTLALSEKEKIEKKVGLSS----AGGNKEEKVIVSYWGIQPSKIT 83
Query: 97 REDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYG 156
++DGTEW WNCF PW TY+ DLSIDL KH P FLDK+A+ TVK+LR PTDVFFQRRYG
Sbjct: 84 KKDGTEWKWNCFSPWGTYKADLSIDLEKHMPPTTFLDKMAFWTVKVLRYPTDVFFQRRYG 143
Query: 157 CRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPK 216
CRAMMLETVAAVPGMV GMLLH +SLR+F+ SGGW KALLEEAENERMHLMT +E+ +PK
Sbjct: 144 CRAMMLETVAAVPGMVAGMLLHCKSLRRFEHSGGWFKALLEEAENERMHLMTFMEVAKPK 203
Query: 217 WYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVP 276
WYER LV+TVQGVFFNA+F+ YLLSPK AHR+ GYLEEEAIHSYTE+LK+++ G IENVP
Sbjct: 204 WYERALVITVQGVFFNAYFLGYLLSPKFAHRMFGYLEEEAIHSYTEFLKELDKGNIENVP 263
Query: 277 APAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
APAIAIDYW+LP +TL+DV+ V+RADEAHHRDVNHFASDIH+ G+ELREA AP+GYH
Sbjct: 264 APAIAIDYWQLPPGSTLRDVVMVVRADEAHHRDVNHFASDIHYQGRELREAAAPIGYH 321
>AOX1_SAUGU (P22185) Alternative oxidase, mitochondrial precursor
(EC 1.-.-.-)
Length = 349
Score = 419 bits (1077), Expect = e-117
Identities = 197/288 (68%), Positives = 238/288 (82%), Gaps = 2/288 (0%)
Query: 49 RMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSV--VVSSYWGISRPKITREDGTEWPWN 106
R S+ +AP ++ +EKA + + ++ G+ V SYW + K+++EDG+EW W
Sbjct: 62 RHASTLSAPAQDGGKEKAAGTAGKVPPGEDGGAEKEAVVSYWAVPPSKVSKEDGSEWRWT 121
Query: 107 CFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVA 166
CF PWETY+ DLSIDL KHHVP LDK+A RTVK LR PTD+FFQRRY CRAMMLETVA
Sbjct: 122 CFRPWETYQADLSIDLHKHHVPTTILDKLALRTVKALRWPTDIFFQRRYACRAMMLETVA 181
Query: 167 AVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTV 226
AVPGMVGG+LLHL+SLR+F+ SGGWI+ALLEEAENERMHLMT +E+ QP+WYER LVL V
Sbjct: 182 AVPGMVGGVLLHLKSLRRFEHSGGWIRALLEEAENERMHLMTFMEVAQPRWYERALVLAV 241
Query: 227 QGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWR 286
QGVFFNA+F+ YLLSPK AHRVVGYLEEEAIHSYTE+LKDI+SGAI++ PAPAIA+DYWR
Sbjct: 242 QGVFFNAYFLGYLLSPKFAHRVVGYLEEEAIHSYTEFLKDIDSGAIQDCPAPAIALDYWR 301
Query: 287 LPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
LP+ +TL+DV+TV+RADEAHHRDVNHFASD+H+ EL+ PAPLGYH
Sbjct: 302 LPQGSTLRDVVTVVRADEAHHRDVNHFASDVHYQDLELKTTPAPLGYH 349
>AOX_CRYNV (Q8NKE2) Alternative oxidase, mitochondrial precursor (EC
1.-.-.-)
Length = 401
Score = 211 bits (536), Expect = 3e-54
Identities = 125/301 (41%), Positives = 175/301 (57%), Gaps = 43/301 (14%)
Query: 58 EEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRP--------KITREDGTEWPWNCFM 109
++ K+ + + S+R EA+K+ G VV S + P + + T W
Sbjct: 50 DDMKRASLSLQPSVR-EAEKSQGPVVGSEGREVEGPHYQDQVSHNVLSDASTTGAWTMLN 108
Query: 110 PWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLR-------------IPTDVFFQR--- 153
P T + +L+ P F DK A+RTVK LR +P V Q+
Sbjct: 109 PIYTEK-ELNTVQVVGRAPVTFGDKAAHRTVKFLRKCFDLLTGYTPYEVPASVLAQKPIP 167
Query: 154 --------------RYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEA 199
++ R ++LE++A VPGMVGG L HLRS+R ++ GGWI +LLEEA
Sbjct: 168 IAELRSKGKLLSDQKWLFRIILLESIAGVPGMVGGTLRHLRSMRLLKRDGGWIHSLLEEA 227
Query: 200 ENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHS 259
ENERMHL+T + + QP + R LVL QGVF+NAFF+ YL+SP++AHR VG LEEEA+ +
Sbjct: 228 ENERMHLLTFMTIAQPGIFTRALVLAAQGVFYNAFFLTYLISPRIAHRFVGALEEEAVRT 287
Query: 260 YTEYLKDIESGAI---ENVPAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASD 316
YT + D+E+G I +++PAPAIAIDYWRLP ++L DVI +RADEA HR VNH ++
Sbjct: 288 YTHCISDMEAGLIPEWKDMPAPAIAIDYWRLPASSSLLDVIRAVRADEATHRFVNHSLAN 347
Query: 317 I 317
+
Sbjct: 348 L 348
>AOX_AJECA (Q9Y711) Alternative oxidase, mitochondrial precursor (EC
1.-.-.-)
Length = 356
Score = 203 bits (516), Expect = 6e-52
Identities = 106/218 (48%), Positives = 142/218 (64%), Gaps = 25/218 (11%)
Query: 125 HHVPKNFLDKVAYRTVKLLRIPTDV---------------------FFQRRYGCRAMMLE 163
H KN+ D VA TV+ LR TD+ +R++ R + LE
Sbjct: 100 HREAKNWSDWVALGTVRFLRWATDLATGYRHAAPGKQGVEVPEQFQMTERKWVIRFIFLE 159
Query: 164 TVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLV 223
TVA VPGMVGGML HLRSLR+ ++ GWI+ LLEEA NERMHL++ ++L QP W+ R +V
Sbjct: 160 TVAGVPGMVGGMLRHLRSLRRMKRDNGWIETLLEEAYNERMHLLSFLKLAQPGWFMRLMV 219
Query: 224 LTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAI---ENVPAPAI 280
L QGVFFN FF+ YL+SP+ HR VGYLEEEA+ +YT +KD+ESG + N PAP I
Sbjct: 220 LGAQGVFFNGFFISYLISPRTCHRFVGYLEEEAVMTYTHAIKDLESGKLPNWANQPAPDI 279
Query: 281 AIDYWRLPK-DATLKDVITVIRADEAHHRDVNHFASDI 317
A+ YW++P+ T+ D++ IRADEA HR+VNH +++
Sbjct: 280 AVAYWQMPEGKRTILDLLYYIRADEAKHREVNHTLANL 317
>AOX_ASPNG (O74180) Alternative oxidase, mitochondrial precursor (EC
1.-.-.-)
Length = 351
Score = 199 bits (505), Expect = 1e-50
Identities = 100/216 (46%), Positives = 144/216 (66%), Gaps = 22/216 (10%)
Query: 125 HHVPKNFLDKVAYRTVKLLRIPTDV------------------FFQRRYGCRAMMLETVA 166
H KN+ D VA TV++LR D+ ++++ R + LE+VA
Sbjct: 98 HRDAKNWADWVALGTVRMLRWGMDLVTGYRHPPPGREHEARFKMTEQKWLTRFIFLESVA 157
Query: 167 AVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTV 226
VPGMVGGML HLRSLR+ ++ GWI+ LLEEA NERMHL+T ++L +P W+ R +VL
Sbjct: 158 GVPGMVGGMLRHLRSLRRMKRDNGWIETLLEEAYNERMHLLTFLKLAEPGWFMRLMVLGA 217
Query: 227 QGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAI---ENVPAPAIAID 283
QGVFFN FF+ YL+SP++ HR VGYLEEEA+ +YT +K+IE+G++ E AP IA+
Sbjct: 218 QGVFFNGFFLSYLMSPRICHRFVGYLEEEAVITYTRAIKEIEAGSLPAWEKTEAPEIAVQ 277
Query: 284 YWRLPK-DATLKDVITVIRADEAHHRDVNHFASDIH 318
YW++P+ ++KD++ +RADEA HR+VNH +++
Sbjct: 278 YWKMPEGQRSMKDLLLYVRADEAKHREVNHTLGNLN 313
>AOX_EMENI (Q9P959) Alternative oxidase, mitochondrial precursor (EC
1.-.-.-)
Length = 354
Score = 197 bits (502), Expect = 2e-50
Identities = 101/210 (48%), Positives = 140/210 (66%), Gaps = 22/210 (10%)
Query: 125 HHVPKNFLDKVAYRTVKLLRIPTDV------------------FFQRRYGCRAMMLETVA 166
H KN+ D VA +V+LLR D+ ++ + R + LE+VA
Sbjct: 101 HREAKNWSDWVALGSVRLLRWGMDLVTGYKHPAPGQEDIKKFQMTEKEWLRRFVFLESVA 160
Query: 167 AVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTV 226
VPGMVGGML HLRSLR+ ++ GWI+ LLEEA NERMHL+T +++ +P W+ R +VL
Sbjct: 161 GVPGMVGGMLRHLRSLRRMKRDNGWIETLLEEAYNERMHLLTFLKMAEPGWFMRLMVLGA 220
Query: 227 QGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAI---ENVPAPAIAID 283
QGVFFN FF+ YL+SP+ HR VGYLEEEA+ +YT +KD+ESG + E + AP IA+
Sbjct: 221 QGVFFNGFFLSYLISPRTCHRFVGYLEEEAVLTYTRAIKDLESGRLPHWEKLEAPEIAVK 280
Query: 284 YWRLPK-DATLKDVITVIRADEAHHRDVNH 312
YW++P+ + T+KD++ +RADEA HR+VNH
Sbjct: 281 YWKMPEGNRTMKDLLLYVRADEAKHREVNH 310
>AOX_BLUGR (Q8X1N9) Alternative oxidase, mitochondrial precursor (EC
1.-.-.-)
Length = 358
Score = 186 bits (471), Expect = 9e-47
Identities = 91/218 (41%), Positives = 138/218 (62%), Gaps = 30/218 (13%)
Query: 125 HHVPKNFLDKVAYRTVKLLRIPTDV------------------------FFQRRYGCRAM 160
H PK+F D++A V+ LR TD+ +R++ R +
Sbjct: 97 HRTPKDFSDRIALYLVRFLRFSTDLATGYKHDPVTITENGEKVLKKPYRMSERKWLIRMV 156
Query: 161 MLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYER 220
LE+VA VPGMV GML HL SLR+ ++ GWI+ LLEEA NERMHL+T +++ +P W+ +
Sbjct: 157 FLESVAGVPGMVAGMLRHLHSLRRLKRDNGWIETLLEEAYNERMHLLTFLKMAKPGWFMK 216
Query: 221 FLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPA- 279
F+++ QGVFFN+ F+ YL+SP+ HR V YLEEEA+ +Y+ ++DIE+G + +P
Sbjct: 217 FMIIGAQGVFFNSMFLSYLISPRTCHRFVAYLEEEAVLTYSTAIQDIEAGLLPKWTSPEF 276
Query: 280 ----IAIDYWRLPK-DATLKDVITVIRADEAHHRDVNH 312
+A+ YW++P+ + T++D++ IRADEA HR+VNH
Sbjct: 277 RIPDLAVQYWKIPEGNRTMRDLLLYIRADEAKHREVNH 314
>AOX_VENIN (Q9P429) Alternative oxidase, mitochondrial precursor (EC
1.-.-.-)
Length = 361
Score = 185 bits (470), Expect = 1e-46
Identities = 102/217 (47%), Positives = 135/217 (62%), Gaps = 29/217 (13%)
Query: 125 HHVPKNFLDKVAYRTVKLLRIPTDVFF------------------QRRYGC-------RA 159
H +N+ DKVA VKLLR D Q+RYG R
Sbjct: 101 HRDTRNWSDKVALIAVKLLRWGLDTVSGYKHGKAQALHAQDPQEAQKRYGMTGKQYLVRN 160
Query: 160 MMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYE 219
+ LE+VA VPGMV GML HL S+R+ ++ GWI+ LLEE+ NERMHL+ ++L +P W+
Sbjct: 161 VFLESVAGVPGMVAGMLRHLHSMRRMKRDHGWIETLLEESYNERMHLLIFLKLYEPGWFM 220
Query: 220 RFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAI---ENVP 276
R VL QGVFFNA F+ YL+SP+ HR VGYLEEEA+ +YT L D+E+G + E +
Sbjct: 221 RLAVLGAQGVFFNAMFLSYLISPRTCHRFVGYLEEEAVVTYTRELADLEAGKLPEWETLA 280
Query: 277 APAIAIDYWRLPK-DATLKDVITVIRADEAHHRDVNH 312
AP IA+DY+ LP+ T+KD++ +RADEA HR+VNH
Sbjct: 281 APDIAVDYYNLPEGHRTMKDLLLHVRADEAKHREVNH 317
>AOX_PODAN (Q9C206) Alternative oxidase, mitochondrial precursor (EC
1.-.-.-)
Length = 363
Score = 184 bits (467), Expect = 3e-46
Identities = 99/283 (34%), Positives = 162/283 (56%), Gaps = 35/283 (12%)
Query: 66 AEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTKH 125
++ S T + + + + ++ + R+ WP + + E L++ + +H
Sbjct: 49 SQSSSQHTRSFSSTRAAHLKDFFPVKETAYIRKTPPAWPHHGYTEEEM----LAV-VPQH 103
Query: 126 HVPKNFLDKVAYRTVKLLRIPTDV------------------------FFQRRYGCRAMM 161
P + D +A++ V+L R TD+ + ++ R +
Sbjct: 104 RKPGSLSDWLAWKLVRLCRWGTDIATGIKPEQQVDKSNPTTAVAAQKPLTEAQWLVRFIF 163
Query: 162 LETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERF 221
LE++A VPGMV GML HL SLR+ ++ GWI+ LLEE+ NERMHL+T +++ +P W+ +
Sbjct: 164 LESIAGVPGMVAGMLRHLESLRRLKRDNGWIETLLEESYNERMHLLTFMKMCEPGWFMKT 223
Query: 222 LVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAI-----ENVP 276
++L QGVFFNA F+ YL+SP++ HR VGYLEEEA+H+YT +++IE G + N
Sbjct: 224 MILGAQGVFFNAMFLSYLISPRITHRFVGYLEEEAVHTYTRCIREIEQGDLPKWSDPNFQ 283
Query: 277 APAIAIDYWRLPK-DATLKDVITVIRADEAHHRDVNHFASDIH 318
P +A+ YW++P+ T++D+I IRADEA HR VNH S+++
Sbjct: 284 IPDLAVTYWKMPEGKRTMRDLILYIRADEAVHRGVNHTLSNLN 326
>AOX_MONFR (Q96UR9) Alternative oxidase, mitochondrial precursor (EC
1.-.-.-) (MfAOX1)
Length = 358
Score = 182 bits (462), Expect = 1e-45
Identities = 105/259 (40%), Positives = 149/259 (56%), Gaps = 50/259 (19%)
Query: 125 HHVPKNFLDKVAYRTVKLLRIPTDV---------------------------FFQRRYGC 157
H P++F D+VA V+ LR TD +R++
Sbjct: 94 HRKPRDFSDRVALGMVRFLRWCTDFATGYKHNVEAPKTASDSNAVTATKPYQMSERKWLI 153
Query: 158 RAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKW 217
R + LE+VA VPGMV GML HLRSLR ++ GWI+ LLEEA NERMHL+T +++ +P
Sbjct: 154 RYVFLESVAGVPGMVAGMLRHLRSLRGLKRDNGWIETLLEEAYNERMHLLTFLKMYEPGL 213
Query: 218 YERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESG-----AI 272
+ R ++L QGVFFN+FF+ YL SPK HR VGYLEEEA+ +YT ++D+E+G A
Sbjct: 214 FMRTMILGAQGVFFNSFFLCYLFSPKTCHRFVGYLEEEAVLTYTLSIQDLENGHLPKWAD 273
Query: 273 ENVPAPAIAIDYWRLPK-DATLKDVITVIRADEAHHRDVNHFASD----------IHFHG 321
N AP +AI+YW +P+ +++D++ IRADEA HR+VNH + + +G
Sbjct: 274 PNFKAPDLAIEYWGMPEGHRSMRDLLYYIRADEAKHREVNHTLGNLKQDEDPNPFVSVYG 333
Query: 322 KELREAPA-------PLGY 333
KE+ + P PLG+
Sbjct: 334 KEVADKPGKGIESLRPLGW 352
>AOX_TRYBB (Q26710) Alternative oxidase, mitochondrial precursor (EC
1.-.-.-)
Length = 329
Score = 181 bits (458), Expect = 3e-45
Identities = 98/213 (46%), Positives = 126/213 (59%), Gaps = 10/213 (4%)
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGC--------RAMMLETVAA 167
PD+ H P +D +AYR+V+ R D F R+G R + LETVA
Sbjct: 68 PDIENVAITHKKPNGLVDTLAYRSVRTCRWLFDTFSLYRFGSITESKVISRCLFLETVAG 127
Query: 168 VPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQ 227
VPGMVGGML HL SLR + GWI LL EAENERMHLMT +EL QP R ++ Q
Sbjct: 128 VPGMVGGMLRHLSSLRYMTRDKGWINTLLVEAENERMHLMTFIELRQPGLPLRVSIIITQ 187
Query: 228 GVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENV--PAPAIAIDYW 285
+ + V Y++SP+ HR VGYLEEEA+ +YT ++ I+ G + P +A YW
Sbjct: 188 AIMYLFLLVAYVISPRFVHRFVGYLEEEAVITYTGVMRAIDEGRLRPTKNDVPEVARVYW 247
Query: 286 RLPKDATLKDVITVIRADEAHHRDVNHFASDIH 318
L K+AT +D+I VIRADEA HR VNH +D+H
Sbjct: 248 NLSKNATFRDLINVIRADEAEHRVVNHTFADMH 280
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.136 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,750,240
Number of Sequences: 164201
Number of extensions: 1679775
Number of successful extensions: 6512
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 6411
Number of HSP's gapped (non-prelim): 48
length of query: 334
length of database: 59,974,054
effective HSP length: 111
effective length of query: 223
effective length of database: 41,747,743
effective search space: 9309746689
effective search space used: 9309746689
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0182.2


