

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0181.4
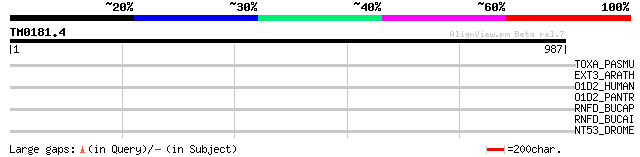
(987 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TOXA_PASMU (P17452) Dermonecrotic toxin (DNT) (PMT) (Mitogenic t... 35 0.76
EXT3_ARATH (Q9FS16) Extensin 3 precursor (AtExt3) (AtExt5) 33 2.9
O1D2_HUMAN (P34982) Olfactory receptor 1D2 (Olfactory receptor-l... 33 4.9
O1D2_PANTR (Q9TUA8) Olfactory receptor 1D2 32 6.4
RNFD_BUCAP (Q8KA19) Electron transport complex protein rnfD 32 8.4
RNFD_BUCAI (P57216) Electron transport complex protein rnfD 32 8.4
NT53_DROME (P82149) Lethal(2)neighbour of tid protein 2 (NOT53) 32 8.4
>TOXA_PASMU (P17452) Dermonecrotic toxin (DNT) (PMT) (Mitogenic
toxin)
Length = 1285
Score = 35.4 bits (80), Expect = 0.76
Identities = 28/113 (24%), Positives = 47/113 (40%), Gaps = 20/113 (17%)
Query: 638 TWHQILSQAPAST-----HSKLTHEESSTTTCSNNLFFPINMYFGPSDLSRLMAHQ---- 688
T+ I S + +S ++ L+ T C N+F + YF DL +A
Sbjct: 82 TYFNIFSSSDSSDGNVFHYNSLSESYRVTDACLMNIF--VERYFDDWDLLNSLASNGIYS 139
Query: 689 ------FSSDHDWGPSSRPM--VKSSTYHLRLIHVLHFVKATWPTHRTYTFMD 733
+ DHD+GP P+ YH R+I + + ++ W + Y FM+
Sbjct: 140 VGKEGAYYPDHDYGPEYNPVWGPNEQIYHSRVIADILYARSVWDEFKKY-FME 191
>EXT3_ARATH (Q9FS16) Extensin 3 precursor (AtExt3) (AtExt5)
Length = 427
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 234 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 262
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 150 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 178
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 318 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 346
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 262 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 290
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 66 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 94
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 206 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 234
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 178 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 206
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 122 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 150
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 94 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 122
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 346 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 374
Score = 33.5 bits (75), Expect = 2.9
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 63 PPPIYHHNPAP-YHNYPSQTQHSALSSPP 90
PPP+ H++P P YH+ P +H SPP
Sbjct: 290 PPPVKHYSPPPVYHSPPPPKKHYVYKSPP 318
Score = 32.7 bits (73), Expect = 4.9
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 6/39 (15%)
Query: 58 SIPPQP-----PPIYHHNPAP-YHNYPSQTQHSALSSPP 90
S PP P PP+ H++P P YH+ P +H SPP
Sbjct: 28 SSPPPPVKHYTPPVKHYSPPPVYHSPPPPKKHYEYKSPP 66
>O1D2_HUMAN (P34982) Olfactory receptor 1D2 (Olfactory receptor-like
protein HGMP07E) (Olfactory receptor 17-4) (OR17-4)
Length = 312
Score = 32.7 bits (73), Expect = 4.9
Identities = 22/69 (31%), Positives = 34/69 (48%), Gaps = 5/69 (7%)
Query: 712 LIHVLHFVKATWPTHRTYTFMDCASNVLLIDGLT--LLTWTLLLAWPHLGCFIFGQPFGF 769
LIH L + T+ R ++ C VLL + + T+L+A GCFIF PFGF
Sbjct: 157 LIHTLLMTRVTFCGSRKIHYIFCEMYVLLRMACSNIQINHTVLIA---TGCFIFLIPFGF 213
Query: 770 ICSKFKFVL 778
+ + ++
Sbjct: 214 VIISYVLII 222
>O1D2_PANTR (Q9TUA8) Olfactory receptor 1D2
Length = 312
Score = 32.3 bits (72), Expect = 6.4
Identities = 22/69 (31%), Positives = 34/69 (48%), Gaps = 5/69 (7%)
Query: 712 LIHVLHFVKATWPTHRTYTFMDCASNVLLIDGLTLL--TWTLLLAWPHLGCFIFGQPFGF 769
LIH L + T+ R ++ C VLL + + T+L+A GCFIF PFGF
Sbjct: 157 LIHTLLMTRVTFCGSRKIHYIFCEMYVLLRMACSNIQTNHTVLIA---TGCFIFLIPFGF 213
Query: 770 ICSKFKFVL 778
+ + ++
Sbjct: 214 VIISYVLII 222
>RNFD_BUCAP (Q8KA19) Electron transport complex protein rnfD
Length = 349
Score = 32.0 bits (71), Expect = 8.4
Identities = 34/167 (20%), Positives = 72/167 (42%), Gaps = 22/167 (13%)
Query: 321 VDSTYALDSLLFKFLAKPGLFIISFDPGICTHTPKLAYGALTKCVAGDFILSHHVFDNSS 380
+ +TY + ++F L ++S PG+CT G L + + F++ +F+
Sbjct: 11 ISNTYDVRKIMF-------LVVLSCIPGLCTEIYFFGCGVLIQTLL--FVIISLLFEII- 60
Query: 381 ERIYKEQQVNIEQFLMHSIMPRTPNLDSLVLASLIGTSLPALLGFIITSFDPGICNAIFS 440
I K ++ NI+ L + S + A L+G S P L + + F I
Sbjct: 61 --ILKMRRKNIKNSLF--------DYSSFLTAVLLGLSTPCALPWWMIIFSCFFAIVISK 110
Query: 441 LSYGDFSKCVFGDSRLSYPGPIMLLTVAKYAANKCDATNALSRFSNL 487
YG + +F + + Y ++L++ + + ++LS ++++
Sbjct: 111 YLYGGLGQNIFNPAMIGY--VVLLISFPVHMTAWNEKNSSLSFYNDI 155
>RNFD_BUCAI (P57216) Electron transport complex protein rnfD
Length = 347
Score = 32.0 bits (71), Expect = 8.4
Identities = 34/136 (25%), Positives = 60/136 (44%), Gaps = 18/136 (13%)
Query: 383 IYKEQQVNIEQFLMHSIMPRTPNLDSLVLAS-LIGTSLPALLGFIITSFDPGICNAIFSL 441
I K + NI+ +L + SLVL S L G S+P LL + +TS +
Sbjct: 56 ILKIRSKNIKNYLQDT---------SLVLTSVLFGVSIPPLLPWWMTSIGLFFAIVVAKH 106
Query: 442 SYGDFSKCVFGDSRLSYPGPIMLLTVAKYAANKCDATNALSRFSNLLPLFDWSYATTLVN 501
YG + +F + + Y ++L++ Y N + +LS F+ D+ + ++
Sbjct: 107 LYGGIGQNIFNPAMVGY--AVLLISFPVYMNNWNERDFSLSFFN------DFKKSAYIIF 158
Query: 502 VPHSSTEVSSKLVSSI 517
+ T VSS ++ I
Sbjct: 159 FKNDITTVSSSYLNII 174
>NT53_DROME (P82149) Lethal(2)neighbour of tid protein 2 (NOT53)
Length = 484
Score = 32.0 bits (71), Expect = 8.4
Identities = 19/67 (28%), Positives = 35/67 (51%), Gaps = 7/67 (10%)
Query: 668 LFFPINMYFGPSDLSRLMAHQFSSDHDWGPSSRPMVKSSTYH-----LRLIHVLHFVKAT 722
L P+ G DL R+ H+++ ++ + SR + ++ T+H L L+ +L F K T
Sbjct: 254 LTHPVEYLRGSFDLGRIFEHKWTVNYRF--LSRDVFENRTFHVSLLGLHLLLLLAFAKPT 311
Query: 723 WPTHRTY 729
W ++Y
Sbjct: 312 WTFFQSY 318
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.137 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 118,677,592
Number of Sequences: 164201
Number of extensions: 5121007
Number of successful extensions: 12058
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 0
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 12016
Number of HSP's gapped (non-prelim): 36
length of query: 987
length of database: 59,974,054
effective HSP length: 120
effective length of query: 867
effective length of database: 40,269,934
effective search space: 34914032778
effective search space used: 34914032778
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0181.4


