

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
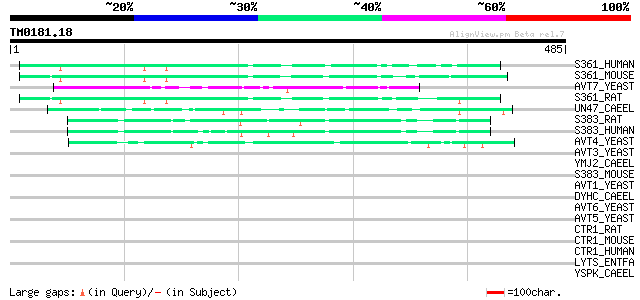
Query= TM0181.18
(485 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
S361_HUMAN (Q7Z2H8) Proton-coupled amino acid transporter 1 (Pro... 64 1e-09
S361_MOUSE (Q8K4D3) Proton-coupled amino acid transporter 1 (Pro... 55 3e-07
AVT7_YEAST (P40501) Vacuolar amino acid transporter 7 54 7e-07
S361_RAT (Q924A5) Proton-coupled amino acid transporter 1 (Proto... 54 1e-06
UN47_CAEEL (P34579) Vesicular GABA transporter (Uncoordinated pr... 49 4e-05
S383_RAT (Q9JHZ9) System N amino acid transporter 1 (SN1) (N-sys... 45 3e-04
S383_HUMAN (Q99624) System N amino acid transporter 1 (SN1) (N-s... 45 5e-04
AVT4_YEAST (P50944) Vacuolar amino acid transporter 4 45 5e-04
AVT3_YEAST (P36062) Vacuolar amino acid transporter 3 44 0.001
YMJ2_CAEEL (P34479) Hypothetical protein F59B2.2 in chromosome III 42 0.004
S383_MOUSE (Q9DCP2) System N amino acid transporter 1 (SN1) (N-s... 42 0.004
AVT1_YEAST (P47082) Vacuolar amino acid transporter 1 41 0.008
DYHC_CAEEL (Q19020) Dynein heavy chain, cytosolic (DYHC) 37 0.11
AVT6_YEAST (P40074) Vacuolar amino acid transporter 6 37 0.11
AVT5_YEAST (P38176) Vacuolar amino acid transporter 5 36 0.25
CTR1_RAT (P30823) High-affinity cationic amino acid transporter-... 35 0.33
CTR1_MOUSE (Q09143) High-affinity cationic amino acid transporte... 35 0.33
CTR1_HUMAN (P30825) High-affinity cationic amino acid transporte... 34 0.74
LYTS_ENTFA (Q82Z75) Sensor protein lytS (EC 2.7.3.-) 33 1.3
YSPK_CAEEL (Q19425) Hypothetical protein F13H10.3 in chromosome IV 33 1.6
>S361_HUMAN (Q7Z2H8) Proton-coupled amino acid transporter 1
(Proton/amino acid transporter 1) (Solute carrier family
36 member 1)
Length = 476
Score = 63.5 bits (153), Expect = 1e-09
Identities = 87/444 (19%), Positives = 177/444 (39%), Gaps = 58/444 (13%)
Query: 9 SKNNHHHQAFDVSLDMQQQGGSKCFDDDGRLKRTG----TVWTASA-HIITAVIGSGVLS 63
+++ H + + DVS + G G +R G T W + H++ IG+G+L
Sbjct: 8 NEDYHDYSSTDVSPEESPSEGLNNLSSPGSYQRFGQSNSTTWFQTLIHLLKGNIGTGLLG 67
Query: 64 LAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVV------- 116
L A+ G V GP +++ +V + +L C + K Y D V
Sbjct: 68 LPLAVKNAGIVMGPISLLIIGIVAVHCMGILVKCAHHFCRRLNKSFVDYGDTVMYGLESS 127
Query: 117 -------HSNMGGIQVKLCGIVQYLN---LFGVAIGYTIASSISMIAIERSNCFHKSEGK 166
H++ G V IV L ++ V + I +NC +
Sbjct: 128 PCSWLRNHAHWGRRVVDFFLIVTQLGFCCVYFVFLADNFKQVIEAANGTTNNCHNNETVI 187
Query: 167 DPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIENG 226
M+ +YM+SF ++L I + L S+LA + + L + + +++
Sbjct: 188 LTPTMDSRLYMLSFLPFLVLLVFIRNLRALSIFSLLANITML----VSLVMIYQFIVQRI 243
Query: 227 AVGGSLTGITVGTVTQTQKVWRTLQAL-GDIAFAYSYSMILIEIQDTVKSPPSESKTMKK 285
L + W+T G F++ +++ +++ +K P +
Sbjct: 244 PDPSHLPLVAP---------WKTYPLFFGTAIFSFEGIGMVLPLENKMKDP----RKFPL 290
Query: 286 ASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAY 345
++ +V+ T+ Y+ GC GY FG + G++ WL + +++ +G +
Sbjct: 291 ILYLGMVIVTILYISLGCLGYLQFGANIQGSITLNL---PNCWLY---QSVKLLYSIGIF 344
Query: 346 QVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITTLISML 405
Y+ ++ Y + FV++ P ++LF RT+ V +T ++++L
Sbjct: 345 FTYA------LQFYVPAEIIIPFFVSRA---PEHCELVVDLF---VRTVLVCLTCILAIL 392
Query: 406 LPFFNDIVGLLGALGFWPLTVYFP 429
+P + ++ L+G++ L + P
Sbjct: 393 IPRLDLVISLVGSVSSSALALIIP 416
>S361_MOUSE (Q8K4D3) Proton-coupled amino acid transporter 1
(Proton/amino acid transporter 1) (Solute carrier family
36 member 1)
Length = 475
Score = 55.5 bits (132), Expect = 3e-07
Identities = 86/450 (19%), Positives = 177/450 (39%), Gaps = 59/450 (13%)
Query: 9 SKNNHHHQAFDVSLDMQQQGGSKCFDDDGRLKRTG-----TVWTASAHIITAVIGSGVLS 63
+++ H + + DVS + G F G +R G T + H++ IG+G+L
Sbjct: 8 NEDYHDYSSTDVSPEESPSEGLGSFSP-GSYQRLGENSSMTWFQTLIHLLKGNIGTGLLG 66
Query: 64 LAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVV------- 116
L A+ G + GP +++ +V + +L C + K Y D V
Sbjct: 67 LPLAVKNAGLLLGPLSLLVIGIVAVHCMGILVKCAHHLCRRLNKPFLDYGDTVMYGLECS 126
Query: 117 -------HSNMGGIQVKLCGIVQYLN---LFGVAIGYTIASSISMIAIERSNCFHKSEGK 166
HS+ G V IV L ++ V + I +NC +
Sbjct: 127 PSTWVRNHSHWGRRIVDFFLIVTQLGFCCVYFVFLADNFKQVIEAANGTTTNCNNNVTVI 186
Query: 167 DPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIENG 226
M+ +YM+SF ++LS I + L S+LA + F + L + + +++
Sbjct: 187 PTPTMDSRLYMLSFLPFLVLLSFIRNLRVLSIFSLLANISMF----VSLIMIYQFIVQRI 242
Query: 227 AVGGSLTGITVGTVTQTQKVWRTLQA-LGDIAFAYSYSMILIEIQDTVKSPPSESKTMKK 285
L + W+T G FA+ +++ +++ +K +S+
Sbjct: 243 PDPSHLPLVA---------PWKTYPLFFGTAIFAFEGIGVVLPLENKMK----DSQKFPL 289
Query: 286 ASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAY 345
++ + + T+ Y+ G GY FG + G++ WL
Sbjct: 290 ILYLGMAIITVLYISLGSLGYLQFGANIKGSITLNL---PNCWLYQSVKL---------- 336
Query: 346 QVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRTIFVIITTLISML 405
+YS +F YA + + ++ + + + +++ + L RT V +T ++++L
Sbjct: 337 -LYSIGIFF---TYALQFYVAAEIIIPAIVSRVPEHFEL-MVDLCVRTAMVCVTCVLAIL 391
Query: 406 LPFFNDIVGLLGALGFWPLTVYFPVEMYII 435
+P + ++ L+G++ L + P + ++
Sbjct: 392 IPRLDLVISLVGSVSSSALALIIPPLLEVV 421
>AVT7_YEAST (P40501) Vacuolar amino acid transporter 7
Length = 490
Score = 54.3 bits (129), Expect = 7e-07
Identities = 71/322 (22%), Positives = 139/322 (43%), Gaps = 29/322 (9%)
Query: 39 LKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACY 98
++ T + +++A+++ ++G+G L++ ++ G + G + +L ++ + +L C
Sbjct: 1 MEATSSALSSTANLVKTIVGAGTLAIPYSFKSDGVLVGVILTLLAAVTSGLGLFVLSKCS 60
Query: 99 RNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSN 158
+ +T + + + I L IVQ FGV + Y + +
Sbjct: 61 KTLINPRNSSFFTLCMLTYPTLAPI-FDLAMIVQ---CFGVGLSYLVLIG---------D 107
Query: 159 CFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLG 218
F G + N ++I+ ++ I L + DQL + SIL + + Y +I +
Sbjct: 108 LFPGLFGGER-----NYWIIASAVIIIPLCLVKKLDQLKYSSILG-LFALAYISILVFSH 161
Query: 219 FGKVIENGAVGGSLTGITVGTVT--QTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSP 276
F V E G G LT I + + L I FA++ SM L + + +K
Sbjct: 162 F--VFELGK--GELTNILRNDICWWKIHDFKGLLSTFSIIIFAFTGSMNLFPMINELKDN 217
Query: 277 PSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAA 336
E+ T + IS+ +T +++ G GY FG+ + GNL+ + N W++ I
Sbjct: 218 SMENITFVINNSISL--STALFLIVGLSGYLTFGNETLGNLMLNYD-PNSIWIV-IGKFC 273
Query: 337 IVIHLVGAYQVYSQPLFAFVEN 358
+ L+ ++ + PL V N
Sbjct: 274 LGSMLILSFPLLFHPLRIAVNN 295
>S361_RAT (Q924A5) Proton-coupled amino acid transporter 1
(Proton/amino acid transporter 1) (Solute carrier family
36 member 1) (Lysosomal amino acid transporter 1)
(LYAAT-1) (Neutral amino acid/proton symporter)
Length = 475
Score = 53.5 bits (127), Expect = 1e-06
Identities = 86/448 (19%), Positives = 171/448 (37%), Gaps = 67/448 (14%)
Query: 9 SKNNHHHQAFDVSLDMQQQGGSKCFDDDGRLKRTG-----TVWTASAHIITAVIGSGVLS 63
+++ H + + DVS + G F G +R G T + H++ IG+G+L
Sbjct: 8 NEDYHDYSSTDVSPEESPSEGLGSFSP-GSYQRLGENSSMTWFQTLIHLLKGNIGTGLLG 66
Query: 64 LAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTYMDVV------- 116
L A+ G + GP +++ +V + +L C + K Y D V
Sbjct: 67 LPLAVKNAGLLLGPLSLLVIGIVAVHCMGILVKCAHHLCRRLNKPFLDYGDTVMYGLECS 126
Query: 117 -------HSNMGGIQVKLCGIVQYLN---LFGVAIGYTIASSISMIAIERSNCFHKSEGK 166
HS+ G V +V L ++ V + I +NC +
Sbjct: 127 PSTWIRNHSHWGRRIVDFFLVVTQLGFCCVYFVFLADNFKQVIEAANGTTTNCNNNETVI 186
Query: 167 DPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIENG 226
M+ +YM++F ++LS I + L S+LA + F + L + + +++
Sbjct: 187 LTPTMDSRLYMLTFLPFLVLLSFIRNLRILSIFSLLANISMF----VSLIMIYQFIVQRI 242
Query: 227 AVGGSLTGITVGTVTQTQKVWRTLQAL-GDIAFAYSYSMILIEIQDTVKSPPSESKTMKK 285
L + W+T G FA+ +++ +++ +K +S+
Sbjct: 243 PDPSHLPLVAP---------WKTYPLFFGTAIFAFEGIGVVLPLENKMK----DSQKFPL 289
Query: 286 ASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAY 345
++ + + T+ Y+ G GY FG G++ WL +++ +G +
Sbjct: 290 ILYLGMAIITVLYISLGSLGYLQFGADIKGSITLNLP---NCWLYQSVK---LLYSIGIF 343
Query: 346 QVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVW----RTIFVIITTL 401
Y+ + E + IP + F LV RT V +T +
Sbjct: 344 FTYALQFYVAAE----------------IIIPAIVSRVPERFELVVDLSARTAMVCVTCV 387
Query: 402 ISMLLPFFNDIVGLLGALGFWPLTVYFP 429
+++L+P + ++ L+G++ L + P
Sbjct: 388 LAVLIPRLDLVISLVGSVSSSALALIIP 415
>UN47_CAEEL (P34579) Vesicular GABA transporter (Uncoordinated
protein 47) (Protein unc-47)
Length = 486
Score = 48.5 bits (114), Expect = 4e-05
Identities = 87/423 (20%), Positives = 170/423 (39%), Gaps = 72/423 (17%)
Query: 34 DDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSIL 93
D G + A+ ++ A+ G ++ L A+ GW + A M+ + V Y+T +L
Sbjct: 80 DGHGEASEPISALQAAWNVTNAIQGMFIVGLPIAVKVGGWWSIGA-MVGVAYVCYWTGVL 138
Query: 94 LCAC-YRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMI 152
L C Y NG K+ TY ++ G K Q L I Y + ++ +
Sbjct: 139 LIECLYENGV----KKRKTYREIADFYKPGFG-KWVLAAQLTELLSTCIIYLVLAADLL- 192
Query: 153 AIERSNCFHKSEGKDPCHMNGNIYMISFGLVEI--VLSQIPDFDQLWWLSI--------L 202
+CF S K M + +++ ++ ++S++ F+ + L + L
Sbjct: 193 ----QSCF-PSVDKAGWMMITSASLLTCSFLDDLQIVSRLSFFNAISHLIVNLIMVLYCL 247
Query: 203 AAVMSFTYSTIGLGLGFGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSY 262
+ V +++STI L T+ T+ +G + F Y+
Sbjct: 248 SFVSQWSFSTITFSLNIN---------------TLPTI------------VGMVVFGYTS 280
Query: 263 SMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFG 322
+ L ++ +K+P ++ M K S I+ V F ++ G G+ FG+ + +
Sbjct: 281 HIFLPNLEGNMKNP-AQFNVMLKWSHIAAAV---FKVVFGMLGFLTFGELTQEEISNSLP 336
Query: 323 FYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAY 382
+ L+++ I +V A Y P YAA + ++ Q P + Y
Sbjct: 337 NQSFKILVNL------ILVVKALLSYPLPF------YAAVQLLKNNLFLGYPQTPFTSCY 384
Query: 383 KINLFRLVW----RTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFPV--EMYIIQ 436
+ W R I V+ T +++ +P+ +++GL+G + L+ +P +YI +
Sbjct: 385 SPDKSLREWAVTLRIILVLFTLFVALSVPYLVELMGLVGNITGTMLSFIWPALFHLYIKE 444
Query: 437 KRI 439
K +
Sbjct: 445 KTL 447
>S383_RAT (Q9JHZ9) System N amino acid transporter 1 (SN1) (N-system
amino acid transporter 1) (Solute carrier family 38,
member 3)
Length = 504
Score = 45.4 bits (106), Expect = 3e-04
Identities = 83/395 (21%), Positives = 156/395 (39%), Gaps = 56/395 (14%)
Query: 51 HIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNY 110
++ A++GSG+L LA+A+A G + ++ +L++ Y+ LL + G R Y
Sbjct: 73 NLSNAIMGSGILGLAYAMANTGIILFLFLLTAVALLSSYSIHLLL----KSSGIVGIRAY 128
Query: 111 TYMDV-VHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKSEGKDPC 169
+ G + L +Q + + Y I S + ++ F E P
Sbjct: 129 EQLGYRAFGTPGKLAAALAITLQNIGAMSSYL-YIIKSELPLVI----QTFLNLEKPTPV 183
Query: 170 -HMNGNIYMISFGLVEIV-LSQIPDFDQLWWLS------ILAAVMSFTYSTIGLGLGFGK 221
+M+GN +I ++ I+ L+ + L + S ++ +++ Y +
Sbjct: 184 WYMDGNYLVILVSVIIILPLALMRQLGYLGYSSGFSLSCMVFFLIAVIYKKFQVPCPLAH 243
Query: 222 VIENGAVGGSLTGITVGTVTQTQKVWRTLQA----------------LGDIAFAYSYSMI 265
+ N A G + V +Q Q T +A + +AFA+
Sbjct: 244 NLVN-ATGNFSHMVVVEEKSQLQSEPDTAEAFCTPSYFTLNSQTAYTIPIMAFAFVCHPE 302
Query: 266 LIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYN 325
++ I +K P + M+ S +S+ V + Y L FGY F D LL + +
Sbjct: 303 VLPIYTELKDP--SKRKMQHISNLSIAVMYVMYFLAALFGYLTFYDGVESELLHTYSKVD 360
Query: 326 PFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKIN 385
PF ++ + V+ V + P+ F V + +Q LF + +
Sbjct: 361 PFDVLILCVRVAVLIAV----TLTVPIVLFP-------------VRRAIQQMLFQNQEFS 403
Query: 386 LFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALG 420
R V I + T I++L+ F +I+G+ G +G
Sbjct: 404 WLRHV--LIATGLLTCINLLVIFAPNILGIFGIIG 436
>S383_HUMAN (Q99624) System N amino acid transporter 1 (SN1)
(N-system amino acid transporter 1) (Solute carrier
family 38, member 3)
Length = 504
Score = 44.7 bits (104), Expect = 5e-04
Identities = 87/394 (22%), Positives = 153/394 (38%), Gaps = 55/394 (13%)
Query: 51 HIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNY 110
++ A++GSG+L LA+A+A G + ++ +L++ Y+ LL V G R Y
Sbjct: 74 NLSNAIMGSGILGLAYAMANTGIILFLFLLTAVALLSSYSIHLLL----KSSGVVGIRAY 129
Query: 111 TYMDV-VHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAIERSNCFHKSEGKDPC 169
+ G + L +Q + + Y I S + ++ N K+
Sbjct: 130 EQLGYRAFGTPGKLAAALAITLQNIGAMSSYL-YIIKSELPLVIQTFLNLEEKTSD---W 185
Query: 170 HMNGNIYMISFGLVEIVLSQIPDFDQLWWLSI-----LAAVMSFTYSTIGLGLGFGKVIE 224
+MNGN Y++ V I+L + QL +L L+ ++ F + I +
Sbjct: 186 YMNGN-YLVILVSVTIILP-LALMRQLGYLGYSSGFSLSCMVFFLIAVIYKKFHVPCPLP 243
Query: 225 ---NGAVGGSLTGITVGTVTQTQKV---------------WRTLQALGDIAFAYSYSMIL 266
N G V Q Q +T + +AFA+ +
Sbjct: 244 PNFNNTTGNFSHVEIVKEKVQLQVEPEASAFCTPSYFTLNSQTAYTIPIMAFAFVCHPEV 303
Query: 267 IEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNP 326
+ I +K P K M+ S +S+ V + Y L FGY F + LL + +P
Sbjct: 304 LPIYTELKDP--SKKKMQHISNLSIAVMYIMYFLAALFGYLTFYNGVESELLHTYSKVDP 361
Query: 327 FWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINL 386
F ++ + V+ V + P+ F V + +Q LF + +
Sbjct: 362 FDVLILCVRVAVLTAV----TLTVPIVLFP-------------VRRAIQQMLFPNQEFSW 404
Query: 387 FRLVWRTIFVIITTLISMLLPFFNDIVGLLGALG 420
R V I V + T I++L+ F +I+G+ G +G
Sbjct: 405 LRHV--LIAVGLLTCINLLVIFAPNILGIFGVIG 436
>AVT4_YEAST (P50944) Vacuolar amino acid transporter 4
Length = 713
Score = 44.7 bits (104), Expect = 5e-04
Identities = 89/408 (21%), Positives = 162/408 (38%), Gaps = 53/408 (12%)
Query: 52 IITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYT 111
++ + IG+GVL L A G +++ F + +Y+ +L V K +
Sbjct: 306 LLKSFIGTGVLFLPNAFHNGGLFFSVSMLAFFGIYSYWCYYIL---------VQAKSSCG 356
Query: 112 YMDVVHSNMGGIQVKLCG-IVQYLNLFGVAIGYTIASSISMIAIERS------NCFHKSE 164
S+ G I +KL G ++ + LF + I S MI ++ N FH
Sbjct: 357 V-----SSFGDIGLKLYGPWMRIIILFSLVITQVGFSGAYMIFTAKNLQAFLDNVFH--V 409
Query: 165 GKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIE 224
G P + M+ ++ I LS I + +L S+LA I GL +
Sbjct: 410 GVLPL----SYLMVFQTIIFIPLSFIRNISKLSLPSLLANFF------IMAGLVIVIIFT 459
Query: 225 NGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMK 284
+ L G V R +G FA+ ++I +QD++++P +
Sbjct: 460 AKRLFFDLMGTPAMGVVYGLNADRWTLFIGTAIFAFEGIGLIIPVQDSMRNPEKFPLVLA 519
Query: 285 KASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGA 344
+ ++ T+ ++ GY A+G + +L N F + +I I L
Sbjct: 520 ----LVILTATILFISIATLGYLAYGSNVQTVILLNLPQSNIFVNLIQLFYSIAIMLSTP 575
Query: 345 YQVYSQPLFAFVENYAAEKF-----PDSDFVTKTVQIPLFNAYKINLFRLVWRTIFV--I 397
Q++ P +EN KF D T+ P N+ K+N +++ W F+ I
Sbjct: 576 LQLF--PAIKIIENKFFPKFTKIYVKHDDLTTRVELRP--NSGKLN-WKIKWLKNFIRSI 630
Query: 398 ITTLISMLLPFFND----IVGLLGALGFWPLTVYFPVEMYIIQKRIPK 441
I ++ + F +D V ++G+L PL +P +++ +P+
Sbjct: 631 IVIIVVSIAYFGSDNLDKFVSVIGSLACIPLVYIYPSMLHLRGNSLPE 678
>AVT3_YEAST (P36062) Vacuolar amino acid transporter 3
Length = 692
Score = 43.9 bits (102), Expect = 0.001
Identities = 83/397 (20%), Positives = 164/397 (40%), Gaps = 40/397 (10%)
Query: 35 DDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILL 94
+ GR + A ++ + +G+GVL L A GW ++ +L++Y + L
Sbjct: 289 EHGRHPHKSSTVKAVLLLLKSFVGTGVLFLPKAFHNGGWGFSALCLLSCALISYGCFVSL 348
Query: 95 CACYRNGDPVNGKRNYTYMDVVHSNMGGIQVKLCGIVQY-LNLFGVAIGYTIASSISMIA 153
D V G Y M + + G ++K + L+ G + YT+ ++ + +
Sbjct: 349 IT---TKDKV-GVDGYGDMGRI---LYGPKMKFAILSSIALSQIGFSAAYTVFTATN-LQ 400
Query: 154 IERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTI 213
+ N FH G ++ Y+ + L+ + LS + +L +++A + I
Sbjct: 401 VFSENFFHLKPGS----ISLATYIFAQVLIFVPLSLTRNIAKLSGTALIADLF------I 450
Query: 214 GLGLGFGKVIENGAVGGSLTGITVGTVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTV 273
LGL + V + ++ G+ T+ K +L +G F + +LI IQ+++
Sbjct: 451 LLGLVYVYVYSIYYI--AVNGVASDTMLMFNKADWSL-FIGTAIFTFEGIGLLIPIQESM 507
Query: 274 KSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIA 333
K P K + + + + + ++ CG YAAFG +L F + L
Sbjct: 508 KHP----KHFRPSLSAVMCIVAVIFISCGLLCYAAFGSDVKTVVLLNFPQDTSYTLTVQL 563
Query: 334 NAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTVQIPLFNAYKINLFRLVWRT 393
A+ I L Q++ P +EN+ FP + +N K+ + +R
Sbjct: 564 LYALAILLSTPLQLF--PAIRILENWT---FPSN-------ASGKYNP-KVKWLKNYFRC 610
Query: 394 IFVIITTLISML-LPFFNDIVGLLGALGFWPLTVYFP 429
V++T++++ + + V L+G+ PL +P
Sbjct: 611 AIVVLTSILAWVGANDLDKFVSLVGSFACIPLIYIYP 647
>YMJ2_CAEEL (P34479) Hypothetical protein F59B2.2 in chromosome III
Length = 460
Score = 42.0 bits (97), Expect = 0.004
Identities = 86/415 (20%), Positives = 166/415 (39%), Gaps = 71/415 (17%)
Query: 41 RTGTVWTASAHIIT---AVIGSGVLSL--AWAIAQLGWVAGPAVMILFSLVTYYTSILLC 95
R+G V T + ++T ++ +G SL AW + L WV+ ++ L Y IL+
Sbjct: 32 RSGDVITPTRAVLTLSKSMFNAGCFSLPYAWKLGGL-WVSFVMSFVIAGLNWYGNHILVR 90
Query: 96 ACYRNGDPVN---------GKRNYTYMDV--VHSNMGGIQ--VKLCGIVQYLNLFGVAIG 142
A + K+ Y D+ + +N + V + + L + VAI
Sbjct: 91 ASQHLAKKSDRSALDYGHFAKKVCDYSDIRFLRNNSKAVMYFVNVTILFYQLGMCSVAIL 150
Query: 143 YTIASSISMIAIERSNCFHKSEGKDPCHMNGNIYMISFGLVEIVLSQIPDFDQLWWLSIL 202
+ + ++++ H+ I M + L I+L+ + F ++ +S
Sbjct: 151 FISDNLVNLVGDHLGGTRHQQM----------ILMATVSLFFILLTNM--FTEMRIVSFF 198
Query: 203 AAVMSFTYSTIGLGLGFGKVIENGAVGGSLTGIT--VGTVTQTQKVWRTLQALGDIAFAY 260
A V S + IG + ++ L T GT+T +G +A+
Sbjct: 199 ALVSS-VFFVIGAAVIMQYTVQQPNQWDKLPAATNFTGTITM----------IGMSMYAF 247
Query: 261 SYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAFGDSSPGNLLTG 320
+++ I++ + +P + S ++++ T F G FGY FGDS + T
Sbjct: 248 EGQTMILPIENKLDNPAAFLAPFGVLS-TTMIICTAFMTALGFFGYTGFGDSIAPTITTN 306
Query: 321 F---GFY---NPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDSDFVTKTV 374
G Y N F ++ +++ + + Y VY F + A +FP+
Sbjct: 307 VPKEGLYSTVNVFLMLQ----SLLGNSIAMYVVYDMFFNGFRRKFGA-RFPN-------- 353
Query: 375 QIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALGFWPLTVYFP 429
+P + + K FR+ W V++T L+++L+P ++ L+G + FP
Sbjct: 354 -VPKWLSDK--GFRVFW----VLVTYLMAVLIPKLEIMIPLVGVTSGALCALIFP 401
>S383_MOUSE (Q9DCP2) System N amino acid transporter 1 (SN1)
(N-system amino acid transporter 1) (Solute carrier
family 38, member 3) (mNAT)
Length = 505
Score = 42.0 bits (97), Expect = 0.004
Identities = 41/173 (23%), Positives = 73/173 (41%), Gaps = 21/173 (12%)
Query: 248 RTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYA 307
+T + +AFA+ ++ I +K P + M+ S +S+ V + Y L FGY
Sbjct: 286 QTAYTIPIMAFAFVCHPEVLPIYTELKDP--SKRKMQHISNLSIAVMYVMYFLAALFGYL 343
Query: 308 AFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFVENYAAEKFPDS 367
F D LL + +PF ++ + V+ V + P+ F
Sbjct: 344 TFYDGVESELLHTYSKVDPFDVLILCVRVAVLIAV----TLTVPIVLFP----------- 388
Query: 368 DFVTKTVQIPLFNAYKINLFRLVWRTIFVIITTLISMLLPFFNDIVGLLGALG 420
V + +Q LF + + R V I + T I++L+ F +I+G+ G +G
Sbjct: 389 --VRRAIQQMLFQNQEFSWLRHV--LIATGLLTCINLLVIFAPNILGIFGIIG 437
>AVT1_YEAST (P47082) Vacuolar amino acid transporter 1
Length = 602
Score = 40.8 bits (94), Expect = 0.008
Identities = 29/102 (28%), Positives = 49/102 (47%), Gaps = 5/102 (4%)
Query: 53 ITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILLCACYRNGDPVNGKRNYTY 112
I +IG G+L+L + GWV G ++ +F+L T+ T+ LL C + DP +Y
Sbjct: 218 INVLIGIGLLALPLGLKYAGWVIGLTMLAIFALATFCTAELLSRCL-DTDPT----LISY 272
Query: 113 MDVVHSNMGGIQVKLCGIVQYLNLFGVAIGYTIASSISMIAI 154
D+ ++ G L + L+L G + I S+ A+
Sbjct: 273 ADLGYAAFGTKGRALISALFTLDLLGSGVSLVILFGDSLNAL 314
>DYHC_CAEEL (Q19020) Dynein heavy chain, cytosolic (DYHC)
Length = 4568
Score = 37.0 bits (84), Expect = 0.11
Identities = 21/58 (36%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Query: 241 TQTQKVWRTLQALGDIAFAYSYSM-ILIEIQDTVKSPPSESKTMKKASFISVVVTTLF 297
T ++ TLQ L +I F Y YS+ L+EI V P S T A + ++ T+LF
Sbjct: 3749 TACSHIYHTLQQLNEIHFLYHYSLDFLVEIFTHVLKTPELSSTTDYAKRLRIITTSLF 3806
>AVT6_YEAST (P40074) Vacuolar amino acid transporter 6
Length = 448
Score = 37.0 bits (84), Expect = 0.11
Identities = 28/107 (26%), Positives = 48/107 (44%), Gaps = 5/107 (4%)
Query: 250 LQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYAAF 309
L L FAY+ + I + +S S + + K I++ + + Y+ GC GY F
Sbjct: 196 LNTLPIFVFAYTCHHNMFSIINEQRS--SRFEHVMKIPLIAISLALILYIAIGCAGYLTF 253
Query: 310 GDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQPLFAFV 356
GD+ GN++ Y I AIV+ ++ A+ + P A +
Sbjct: 254 GDNIIGNIIM---LYPQAVSSTIGRIAIVLLVMLAFPLQCHPARASI 297
>AVT5_YEAST (P38176) Vacuolar amino acid transporter 5
Length = 509
Score = 35.8 bits (81), Expect = 0.25
Identities = 30/127 (23%), Positives = 57/127 (44%), Gaps = 16/127 (12%)
Query: 250 LQALGDIAFAYS--YSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFYMLCGCFGYA 307
L L FAY+ ++M + + KS K +++ ++V+ Y++ G GY
Sbjct: 256 LTTLPIFVFAYTCHHNMFSVINEQVDKS----FKVIRRIPIFAIVLAYFLYIIIGGTGYM 311
Query: 308 AFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAYQVYSQP-------LFAFVENYA 360
FG++ GN+LT Y I A+++ ++ A+ + P + F+EN+
Sbjct: 312 TFGENIVGNILT---LYPNSISTTIGRLAMLLLVMLAFPLQCHPCRSSVKNIIIFIENFR 368
Query: 361 AEKFPDS 367
K D+
Sbjct: 369 KGKLYDN 375
>CTR1_RAT (P30823) High-affinity cationic amino acid transporter-1
(CAT-1) (CAT1) (System Y+ basic amino acid transporter)
(Ecotropic retroviral leukemia receptor) (ERR)
(Ecotropic retrovirus receptor)
Length = 624
Score = 35.4 bits (80), Expect = 0.33
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query: 32 CFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTS 91
C ++ RL R + A + + +G+GV LA A+A+ AGPA++I F L+ S
Sbjct: 21 CSREESRLSRCLNTYDLVALGVGSTLGAGVYVLAGAVAREN--AGPAIVISF-LIAALAS 77
Query: 92 ILLCACY 98
+L CY
Sbjct: 78 VLAGLCY 84
>CTR1_MOUSE (Q09143) High-affinity cationic amino acid
transporter-1 (CAT-1) (CAT1) (System Y+ basic amino
acid transporter) (Ecotropic retroviral leukemia
receptor) (ERR) (Ecotropic retrovirus receptor)
Length = 622
Score = 35.4 bits (80), Expect = 0.33
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query: 32 CFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTS 91
C ++ RL R + A + + +G+GV LA A+A+ AGPA++I F L+ S
Sbjct: 21 CSREESRLSRCLNTYDLVALGVGSTLGAGVYVLAGAVAREN--AGPAIVISF-LIAALAS 77
Query: 92 ILLCACY 98
+L CY
Sbjct: 78 VLAGLCY 84
>CTR1_HUMAN (P30825) High-affinity cationic amino acid
transporter-1 (CAT-1) (CAT1) (System Y+ basic amino
acid transporter) (Ecotropic retroviral leukemia
receptor homolog) (ERR) (Ecotropic retrovirus receptor
homolog)
Length = 629
Score = 34.3 bits (77), Expect = 0.74
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query: 32 CFDDDGRLKRTGTVWTASAHIITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTS 91
C ++ RL R + A + + +G+GV LA A+A+ AGPA++I F L+ S
Sbjct: 21 CSREETRLSRCLNTFDLVALGVGSTLGAGVYVLAGAVAREN--AGPAIVISF-LIAALAS 77
Query: 92 ILLCACY 98
+L CY
Sbjct: 78 VLAGLCY 84
>LYTS_ENTFA (Q82Z75) Sensor protein lytS (EC 2.7.3.-)
Length = 589
Score = 33.5 bits (75), Expect = 1.3
Identities = 43/167 (25%), Positives = 69/167 (40%), Gaps = 23/167 (13%)
Query: 179 SFGLVEIVLSQIPDFDQLWWLSILAAVMSFTYSTIGLGLGFGKVIENGAVGGSLTGITVG 238
+F +EI +QI + L +LS A++ + IG+ +G VGG + G VG
Sbjct: 56 NFTGIEIAKNQIVPNNLLTYLSSNASIANTRTLVIGV---------SGLVGGPIVGSAVG 106
Query: 239 TVTQTQKVWRTLQALGDIAFAYSYSMILIEIQDTVKSPPSESKTMKKASFISVVVTTLFY 298
+ +V +Q G F S+I+ I + S ++ A F S +V
Sbjct: 107 LIAGFHRV---IQGGGHSFFYVPASLIVGLIAGFLGSRMAKQTVFPSAGF-SAIVGACME 162
Query: 299 MLCGCFGYAAFGDSSPGNLLTGFGFYNPFWLIDIANAAIVIHLVGAY 345
M+ F + GD S G L F IA I+++ VG +
Sbjct: 163 MIQMIFIFFFSGDLSDGATLVRF----------IALPMILLNSVGTF 199
>YSPK_CAEEL (Q19425) Hypothetical protein F13H10.3 in chromosome IV
Length = 615
Score = 33.1 bits (74), Expect = 1.6
Identities = 12/43 (27%), Positives = 27/43 (61%)
Query: 52 IITAVIGSGVLSLAWAIAQLGWVAGPAVMILFSLVTYYTSILL 94
I ++G+ +L++ WA+ Q G V G +M+ + + +YT+ ++
Sbjct: 172 IWNTMMGTSLLAMPWALQQAGLVLGIIIMLSMAAICFYTAYIV 214
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.141 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,267,849
Number of Sequences: 164201
Number of extensions: 2331950
Number of successful extensions: 6998
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 6961
Number of HSP's gapped (non-prelim): 49
length of query: 485
length of database: 59,974,054
effective HSP length: 114
effective length of query: 371
effective length of database: 41,255,140
effective search space: 15305656940
effective search space used: 15305656940
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0181.18


