

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
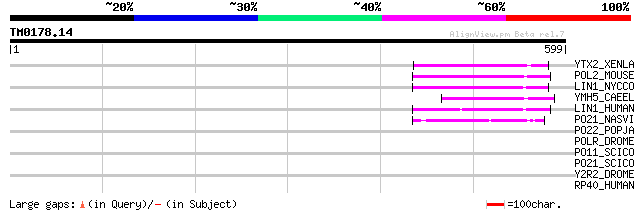
Query= TM0178.14
(599 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 77 1e-13
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 69 3e-11
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 66 3e-10
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 59 3e-08
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 59 5e-08
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 45 7e-04
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 37 0.19
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 36 0.33
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 34 0.96
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 34 1.2
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 33 2.1
RP40_HUMAN (O75818) Ribonuclease P protein subunit p40 (EC 3.1.2... 32 3.6
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 77.4 bits (189), Expect = 1e-13
Identities = 46/146 (31%), Positives = 75/146 (50%), Gaps = 3/146 (2%)
Query: 436 KFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITH 495
K G DGL F++ WDT+ F F G LP V++L+PK + I +
Sbjct: 465 KSPGLDGLTIEFFQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKN 524
Query: 496 YRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRK 555
+RP+S + YK++++ I RLK L ++I +QS + + I DN+ + ++ H R
Sbjct: 525 WRPVSLLSTDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRT 584
Query: 556 GRASKEHVALKLDMSKAYERVEWSFL 581
G + L LD KA++RV+ +L
Sbjct: 585 GLSL---AFLSLDQEKAFDRVDHQYL 607
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 68.9 bits (167), Expect = 3e-11
Identities = 42/150 (28%), Positives = 74/150 (49%), Gaps = 3/150 (2%)
Query: 435 RKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVP-NPESI 493
+K GPDG + FY+ + + + + G LP+ E +TLIPK +P I
Sbjct: 494 KKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGTLPNSFYEATITLIPKPQKDPTKI 553
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
++RPIS N K++++I+ R++ + +I +Q FI NI + H I
Sbjct: 554 ENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYIN 613
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLEK 583
+ K H+ + LD KA+++++ F+ K
Sbjct: 614 KL--KDKNHMIISLDAEKAFDKIQHPFMIK 641
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 65.9 bits (159), Expect = 3e-10
Identities = 40/148 (27%), Positives = 74/148 (49%), Gaps = 3/148 (2%)
Query: 435 RKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKV-PNPESI 493
+K GPDG FY+ + + + ++ G LP+ E +TLIPK +P
Sbjct: 467 KKSPGPDGFTSEFYQTFKEELVPILLNLFQNIEKEGILPNTFYEANITLIPKPGKDPTRK 526
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
+YRPIS N K++++I+ R++ + ++I +Q FI NI + +I
Sbjct: 527 ENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQHIN 586
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFL 581
+ +K+H+ L +D KA++ ++ F+
Sbjct: 587 K--LKNKDHMILSIDAEKAFDNIQHPFM 612
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 59.3 bits (142), Expect = 3e-08
Identities = 34/122 (27%), Positives = 64/122 (51%), Gaps = 3/122 (2%)
Query: 467 FSTGHLPSKINETVVTLIPKVPNPESITHYRPISCCNFTYKVISRIIVTRLKGGLNQLIT 526
FS +P++ V+ IPK NP S ++YRPIS + +++ RII +R++ + L++
Sbjct: 656 FSDSAIPNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLS 715
Query: 527 LNQSAFIGVKMIQDNIMIAQEAFHNILRKGRASKEHVALKLDMSKAYERVEWSFLEKNPP 586
+Q F+ + +++ + +H+IL+ K L D +KA+++V L K
Sbjct: 716 PHQHGFLNFRSCPSSLVRSISLYHSILKN---EKSLDILFFDFAKAFDKVSHPILLKKLA 772
Query: 587 CF 588
F
Sbjct: 773 LF 774
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 58.5 bits (140), Expect = 5e-08
Identities = 38/151 (25%), Positives = 74/151 (48%), Gaps = 5/151 (3%)
Query: 435 RKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI- 493
+K GP+G FY+ + + + + +S G LP+ E + LIPK P ++
Sbjct: 467 KKSPGPEGFTAEFYQRYKEELVPFLLKLFQSIEKEGILPNSFYEASIILIPK-PGRDTTK 525
Query: 494 -THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNI 552
++RPIS N K++++I+ +++ + +LI +Q FI NI + +I
Sbjct: 526 KENFRPISLMNIDAKILNKILANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKSINIIQHI 585
Query: 553 LRKGRASKEHVALKLDMSKAYERVEWSFLEK 583
R H+ + +D KA+++++ F+ K
Sbjct: 586 NR--TKDTNHMIISIDAEKAFDKIQQPFMLK 614
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 44.7 bits (104), Expect = 7e-04
Identities = 35/143 (24%), Positives = 60/143 (41%), Gaps = 9/143 (6%)
Query: 435 RKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESIT 494
R GPDG+ W+++ E G P + ++ LIPK P
Sbjct: 332 RTAAGPDGMT----TTAWNSIDECIKSLFNMIMYHGQCPRRYLDSRTVLIPKEPGTMDPA 387
Query: 495 HYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILR 554
+RP+S + + RI+ R+ G + L+ Q AFI + +N +
Sbjct: 388 CFRPLSIASVALRHFHRILANRI--GEHGLLDTRQRAFIVADGVAENTSLLSAMIKEARM 445
Query: 555 KGRASKEHVALKLDMSKAYERVE 577
K + ++A+ LD+ KA++ VE
Sbjct: 446 KIKGL--YIAI-LDVKKAFDSVE 465
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 36.6 bits (83), Expect = 0.19
Identities = 27/110 (24%), Positives = 55/110 (49%), Gaps = 5/110 (4%)
Query: 470 GHLPSKINETVVTLIPKVPNPESITHYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQ 529
GH+P+ TLIPK + E+ +++RPI+ + +++ RI+ RL+ +
Sbjct: 50 GHVPTPWTAMRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVELHPAQKG 109
Query: 530 SAFIGVKMIQDNIMIAQEAFHNILRKGRASKEHVALKLDMSKAYERVEWS 579
A I ++ ++ + + + R+ R K + + LD+ KA++ V S
Sbjct: 110 YARIDGTLVNSLLL---DTYISSRREQR--KTYNVVSLDVRKAFDTVSHS 154
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 35.8 bits (81), Expect = 0.33
Identities = 33/136 (24%), Positives = 55/136 (40%), Gaps = 8/136 (5%)
Query: 439 GPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITHYRP 498
GPDG+ K+ + + G+LP I IPK + +RP
Sbjct: 361 GPDGITP---KSAREVPSGIMLRIMNLILWCGNLPHSIRLARTVFIPKTVTAKRPQDFRP 417
Query: 499 ISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRKGRA 558
IS + + ++ I+ TRL +N Q F+ DN I + + R+
Sbjct: 418 ISVPSVLVRQLNAILATRLNSSINW--DPRQRGFLPTDGCADNATIVDLVLRHSHKHFRS 475
Query: 559 SKEHVALKLDMSKAYE 574
++A LD+SKA++
Sbjct: 476 C--YIA-NLDVSKAFD 488
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 34.3 bits (77), Expect = 0.96
Identities = 38/152 (25%), Positives = 62/152 (40%), Gaps = 18/152 (11%)
Query: 435 RKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKV-----PN 489
RK GPDG+ G + W + E + + P K + ++ K+ +
Sbjct: 420 RKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIASLVILLKLLDRIRSD 479
Query: 490 PESITHYRPISCCNFTYKVISRIIVTRLKGGLNQL-ITLNQSAFIGVKMIQDNIMIAQEA 548
P S YRPI + KV+ I+V RL L + ++ Q AF K +D A
Sbjct: 480 PGS---YRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKSTED-------A 529
Query: 549 FHNILRKGRAS--KEHVALKLDMSKAYERVEW 578
+ + R S K + L +D A++ + W
Sbjct: 530 WRCVQRHVECSEMKYVIGLNIDFQGAFDNLGW 561
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 33.9 bits (76), Expect = 1.2
Identities = 35/148 (23%), Positives = 62/148 (41%), Gaps = 12/148 (8%)
Query: 436 KFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFST-GHLPSKINETVVTLIPKVPNPESIT 494
K GPDG+ W+ + + + + F G +PS I + PK+
Sbjct: 173 KGAGPDGVT----PRSWNALDDRYKRLLYNIFVFYGRVPSPIKGSRTVFTPKIEGGPDPG 228
Query: 495 HYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILR 554
+RP+S C+ + ++I+ R Q+A++ + + N+ + I
Sbjct: 229 VFRPLSICSVILREFNKILARRFVSCYT--YDERQTAYLPIDGVCINVSMLTAI---IAE 283
Query: 555 KGRASKE-HVALKLDMSKAYERVEWSFL 581
R KE H+A+ LD+ KA+ V S L
Sbjct: 284 AKRLRKELHIAI-LDLVKAFNSVYHSAL 310
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 33.1 bits (74), Expect = 2.1
Identities = 36/149 (24%), Positives = 60/149 (40%), Gaps = 12/149 (8%)
Query: 435 RKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI- 493
R+ G DG+NG K W + E G+ P++ V + K P+ +
Sbjct: 449 RRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVSLLKGPDKDKCE 508
Query: 494 -THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNI 552
+ YR I KV+ I+V R++ L + Q F + ++D A+ ++
Sbjct: 509 PSSYRGICLLPVFGKVLEAIMVNRVREVLPEGCRW-QFGFRQGRCVED-------AWRHV 560
Query: 553 LRKGRASKEHVALK--LDMSKAYERVEWS 579
AS L +D A++ VEWS
Sbjct: 561 KSSVGASAAQYVLGTFVDFKGAFDNVEWS 589
>RP40_HUMAN (O75818) Ribonuclease P protein subunit p40 (EC
3.1.26.5) (RNaseP protein p40) (RNase P subunit 1)
Length = 302
Score = 32.3 bits (72), Expect = 3.6
Identities = 16/28 (57%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query: 563 VALKLDMSKAYERVEWSFLEKNPPCFRF 590
++L LD SK YER+ WSF EK P F F
Sbjct: 93 LSLNLD-SKKYERISWSFKEKKPLKFDF 119
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.350 0.155 0.548
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 60,993,553
Number of Sequences: 164201
Number of extensions: 2245448
Number of successful extensions: 7084
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 7072
Number of HSP's gapped (non-prelim): 13
length of query: 599
length of database: 59,974,054
effective HSP length: 116
effective length of query: 483
effective length of database: 40,926,738
effective search space: 19767614454
effective search space used: 19767614454
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0178.14


