

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.15
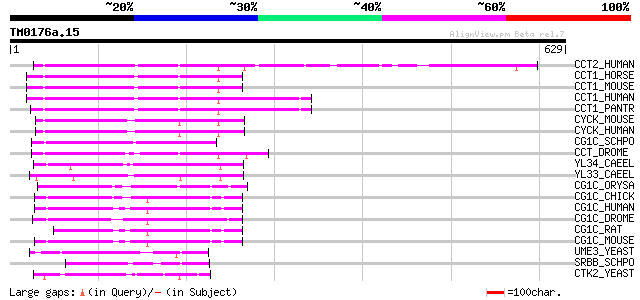
(629 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CCT2_HUMAN (O60583) Cyclin T2 164 6e-40
CCT1_HORSE (Q9XT26) Cyclin T1 (Cyclin T) (CycT1) 156 1e-37
CCT1_MOUSE (Q9QWV9) Cyclin T1 (Cyclin T) (CycT1) 156 2e-37
CCT1_HUMAN (O60563) Cyclin T1 (Cyclin T) (CycT1) 156 2e-37
CCT1_PANTR (Q8HXN7) Cyclin T1 (Cyclin T) (CycT1) 155 3e-37
CYCK_MOUSE (O88874) Cyclin K 152 3e-36
CYCK_HUMAN (O75909) Cyclin K 152 3e-36
CG1C_SCHPO (O74627) Cyclin pch1 (Pombe cyclin c homolog 1) 136 1e-31
CCT_DROME (O96433) Cyclin T 123 1e-27
YL34_CAEEL (P34425) Hypothetical protein F44B9.4 in chromosome III 92 4e-18
YL33_CAEEL (P34424) Hypothetical protein F44B9.3 in chromosome III 89 5e-17
CG1C_ORYSA (P93411) G1/S-specific cyclin C-type 86 4e-16
CG1C_CHICK (P55168) Cyclin C 86 4e-16
CG1C_HUMAN (P24863) Cyclin C 85 7e-16
CG1C_DROME (P25008) G1/S-specific cyclin C 85 7e-16
CG1C_RAT (P39947) Cyclin C 83 2e-15
CG1C_MOUSE (Q62447) Cyclin C 81 7e-15
UME3_YEAST (P47821) RNA polymerase II holoenzyme cyclin-like sub... 65 4e-10
SRBB_SCHPO (O94503) RNA polymerase II holoenzyme cyclin-like sub... 64 1e-09
CTK2_YEAST (P46962) CTD kinase beta subunit (CTD kinase 38 kDa s... 55 7e-07
>CCT2_HUMAN (O60583) Cyclin T2
Length = 730
Score = 164 bits (415), Expect = 6e-40
Identities = 151/597 (25%), Positives = 276/597 (45%), Gaps = 62/597 (10%)
Query: 28 SRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHR 87
SRW+ +R+++E N+PS++ GV+ KE R+ +Q++G RL V Q+TI +AI++ HR
Sbjct: 9 SRWFFTREQLE-NTPSRRCGVEADKELSCRQQAANLIQEMGQRLNVSQLTINTAIVYMHR 67
Query: 88 FFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEV 147
F++ S K ++ II+++ +FLA KVEE R L+ VI V++ +H +P + +
Sbjct: 68 FYMHHSFTKFNKNIISSTALFLAAKVEEQARKLEHVIKVAHACLHPLEPLLDTKC---DA 124
Query: 148 YEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLR- 206
Y QQ + +++ E ++L TLGF+ ++HP+ +V+ + ++ S++ LAQ ++ + L
Sbjct: 125 YLQQTQELVILETIMLQTLGFEITIEHPHTDVVKCTQLVRASKD-LAQTSYFMATNSLHL 183
Query: 207 TSLCLQFKPQHIAAGAIFLAAKFLNVKLE-----KLWWQVFDVTLTAHQLEEVSNQMLEL 261
T+ CLQ+KP IA I LA K+ N ++ K WW+ D T+T L+E++++ L++
Sbjct: 184 TTFCLQYKPTVIACVCIHLACKWSNWEIPVSTDGKHWWEYVDPTVTLELLDELTHEFLQI 243
Query: 262 YEQ-------------NQIAPSNDVEGTAGGGNRATPKASTSNDEAATTNNNLYIRGPSP 308
E+ NQ A V+G TP +S + + +++ +P
Sbjct: 244 LEKTPNRLKKIRNWRANQAARKPKVDGQVS----ETPLLGSSLVQNSILVDSVTGVPTNP 299
Query: 309 RLE-ASKLAISKNVVSSANHVGPVSNHGTNGSTEMIHLVEGDAKGNQHPEQEPPPHKENL 367
+ S A V ++ ++ +H T+ + M+ QE P H
Sbjct: 300 SFQKPSTSAFPAPVPLNSGNISVQDSH-TSDNLSMLATGMPSTSYGLSSHQEWPQH---- 354
Query: 368 QEAQDTVRSRSGYSEEPEINIGRSNMKEDVELNDKHS-SKNLNHRDGTFAPPSLEAIKKI 426
QD+ R+ YS++ E ++ S + + S L+HR + S ++K+
Sbjct: 355 ---QDSARTEQLYSQKQETSLSGSQYNINFQQGPSISLHSGLHHRPDKISDHS--SVKQE 409
Query: 427 DRDKVKAALEKRKKAVGNITKKTELMDDEDLFERVRELEDGIELAAQSEKNKQDDEDLIE 486
K ++ K G I+ ++ + ++ R EK K + DL
Sbjct: 410 YTHKAGSS-----KHHGPISTTPGIIPQKMSLDKYR------------EKRKLETLDLDV 452
Query: 487 RVRELEDGIELAAQSEKNKQDGQKSLSKPVDRSAFENMQQGKHQDHSEE--HLHLVKPSH 544
R + +E + +++ S++ P+ + D E+ L L P
Sbjct: 453 RDHYIAAQVEQQHKQGQSQAASSSSVTSPIKMKIPIANTEKYMADKKEKSGSLKLRIPIP 512
Query: 545 ETDLSAVEEGEVSALDDIDLGRKSSNHK---RKAESSPDKFAEGKKRHNYSFGSTHH 598
TD SA +E + R SS+ + + SSP + K++H S HH
Sbjct: 513 PTDKSASKEELKMKIKVSSSERHSSSDEGSGKSKHSSPHISRDHKEKHKEHPSSRHH 569
>CCT1_HORSE (Q9XT26) Cyclin T1 (Cyclin T) (CycT1)
Length = 727
Score = 156 bits (395), Expect = 1e-37
Identities = 87/251 (34%), Positives = 150/251 (59%), Gaps = 11/251 (4%)
Query: 20 QGCSQENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIA 79
+G + N RWY +R+++E NSPS++ G+D KE R+ LQD+G RL V Q+TI
Sbjct: 2 EGERKNNNKRWYFTREQLE-NSPSRRFGLDPDKELSYRQQAANLLQDMGQRLNVSQLTIN 60
Query: 80 SAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAV 139
+AI++ HRF++ QS + R +A + +FLA KVEE P+ L+ VI V++ +H ++
Sbjct: 61 TAIVYMHRFYMIQSFTQFHRNSVAPAALFLAAKVEEQPKKLEHVIKVAHACLHPQESLPD 120
Query: 140 QRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWN 199
R E Y QQ + +++ E ++L TLGF+ + HP+ +V+ + ++ S++ LAQ ++
Sbjct: 121 TR---SEAYLQQVQDLVILESIILQTLGFELTIDHPHTHVVKCTQLVRASKD-LAQTSYF 176
Query: 200 FVNDGLR-TSLCLQFKPQHIAAGAIFLAAKFLNVKLE-----KLWWQVFDVTLTAHQLEE 253
+ L T+ LQ+ P +A I LA K+ N ++ K WW+ D T+T L+E
Sbjct: 177 MATNSLHLTTFSLQYTPPVVACVCIHLACKWSNWEIPVSTDGKHWWEYVDATVTLELLDE 236
Query: 254 VSNQMLELYEQ 264
++++ L++ E+
Sbjct: 237 LTHEFLQILEK 247
>CCT1_MOUSE (Q9QWV9) Cyclin T1 (Cyclin T) (CycT1)
Length = 724
Score = 156 bits (394), Expect = 2e-37
Identities = 88/251 (35%), Positives = 150/251 (59%), Gaps = 11/251 (4%)
Query: 20 QGCSQENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIA 79
+G + N RWY +R+++E NSPS++ GVD KE R+ LQD+G RL V Q+TI
Sbjct: 2 EGERKNNNKRWYFTREQLE-NSPSRRFGVDSDKELSYRQQAANLLQDVGQRLNVSQLTIN 60
Query: 80 SAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAV 139
+AI++ HRF++ QS + R +A + +FLA KVEE P+ L+ VI V++ +H ++
Sbjct: 61 TAIVYMHRFYMIQSFTQFHRYSMAPAALFLAAKVEEQPKKLEHVIKVAHTCLHPQESLPD 120
Query: 140 QRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWN 199
R E Y QQ + +++ E ++L TLGF+ + HP+ +V+ + ++ S++ LAQ ++
Sbjct: 121 TR---SEAYLQQVQDLVILESIILQTLGFELTIDHPHTHVVKCTQLVRASKD-LAQTSYF 176
Query: 200 FVNDGLR-TSLCLQFKPQHIAAGAIFLAAKFLNVKLE-----KLWWQVFDVTLTAHQLEE 253
+ L T+ LQ+ P +A I LA K+ N ++ K WW+ D T+T L+E
Sbjct: 177 MATNSLHLTTFSLQYTPPVVACVCIHLACKWSNWEIPVSTDGKHWWEYVDATVTLELLDE 236
Query: 254 VSNQMLELYEQ 264
++++ L++ E+
Sbjct: 237 LTHEFLQILEK 247
>CCT1_HUMAN (O60563) Cyclin T1 (Cyclin T) (CycT1)
Length = 726
Score = 156 bits (394), Expect = 2e-37
Identities = 101/331 (30%), Positives = 176/331 (52%), Gaps = 14/331 (4%)
Query: 20 QGCSQENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIA 79
+G + N RWY +R+++E NSPS++ GVD KE R+ LQD+G RL V Q+TI
Sbjct: 2 EGERKNNNKRWYFTREQLE-NSPSRRFGVDPDKELSYRQQAANLLQDMGQRLNVSQLTIN 60
Query: 80 SAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAV 139
+AI++ HRF++ QS + +A + +FLA KVEE P+ L+ VI V++ +H ++
Sbjct: 61 TAIVYMHRFYMIQSFTQFPGNSVAPAALFLAAKVEEQPKKLEHVIKVAHTCLHPQESLPD 120
Query: 140 QRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWN 199
R E Y QQ + +++ E ++L TLGF+ + HP+ +V+ + ++ S++ LAQ ++
Sbjct: 121 TR---SEAYLQQVQDLVILESIILQTLGFELTIDHPHTHVVKCTQLVRASKD-LAQTSYF 176
Query: 200 FVNDGLR-TSLCLQFKPQHIAAGAIFLAAKFLNVKLE-----KLWWQVFDVTLTAHQLEE 253
+ L T+ LQ+ P +A I LA K+ N ++ K WW+ D T+T L+E
Sbjct: 177 MATNSLHLTTFSLQYTPPVVACVCIHLACKWSNWEIPVSTDGKHWWEYVDATVTLELLDE 236
Query: 254 VSNQMLELYEQNQIAPSNDVEGTAGGGNRATPKASTSNDEAAT--TNNNLYIRGPSPRLE 311
++++ L++ E+ A + T DE + T N+ + S
Sbjct: 237 LTHEFLQILEKTPNRLKRIWNWRACEAAKKTKADDRGTDEKTSEQTILNMISQSSSDTTI 296
Query: 312 ASKLAISKNVVSSANHVGPVSNHGTNGSTEM 342
A +++S + S+ + PVS ++ T +
Sbjct: 297 AGLMSMSTSTTSAVPSL-PVSEESSSNLTSV 326
>CCT1_PANTR (Q8HXN7) Cyclin T1 (Cyclin T) (CycT1)
Length = 725
Score = 155 bits (392), Expect = 3e-37
Identities = 100/327 (30%), Positives = 175/327 (52%), Gaps = 14/327 (4%)
Query: 24 QENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAII 83
++N RWY +R+++E NSPS++ GVD KE R+ LQD+G RL V Q+TI +AI+
Sbjct: 5 RKNNKRWYFTREQLE-NSPSRRFGVDPDKELSYRQQAANLLQDMGQRLNVSQLTINTAIV 63
Query: 84 FCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIK 143
+ HRF++ QS + +A + +FLA KVEE P+ L+ VI V++ +H ++ R
Sbjct: 64 YMHRFYMIQSFTQFPGNSVAPAALFLAAKVEEQPKKLEHVIKVAHTCLHPQESLPDTR-- 121
Query: 144 QKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVND 203
E Y QQ + +++ E ++L TLGF+ + HP+ +V+ + ++ S++ LAQ ++ +
Sbjct: 122 -SEAYLQQVQDLVILESIILQTLGFELTIDHPHTHVVKCTQLVRASKD-LAQTSYFMATN 179
Query: 204 GLR-TSLCLQFKPQHIAAGAIFLAAKFLNVKLE-----KLWWQVFDVTLTAHQLEEVSNQ 257
L T+ LQ+ P +A I LA K+ N ++ K WW+ D T+T L+E++++
Sbjct: 180 SLHLTTFSLQYTPPVVACVCIHLACKWSNWEIPVSTDGKHWWEYVDATVTLELLDELTHE 239
Query: 258 MLELYEQNQIAPSNDVEGTAGGGNRATPKASTSNDEAAT--TNNNLYIRGPSPRLEASKL 315
L++ E+ A + T DE + T N+ + S A +
Sbjct: 240 FLQILEKTPNRLKRIWNWRACEAAKKTKADDRGTDEKTSEQTILNMISQSSSDTTIAGLM 299
Query: 316 AISKNVVSSANHVGPVSNHGTNGSTEM 342
++S + S+ + PVS ++ T +
Sbjct: 300 SMSTSTTSAVPSL-PVSEESSSNLTSV 325
>CYCK_MOUSE (O88874) Cyclin K
Length = 554
Score = 152 bits (383), Expect = 3e-36
Identities = 82/248 (33%), Positives = 145/248 (58%), Gaps = 20/248 (8%)
Query: 30 WYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFF 89
WY +K++ ++PS+ +G+D EA R+ ++ D+G RL + T+A+ II+ HRF+
Sbjct: 24 WYWDKKDLA-HTPSQLEGLDPATEARYRREGARFIFDVGTRLGLHYDTLANGIIYFHRFY 82
Query: 90 LRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYE 149
+ S + R + C+FLAGKVEETP+ KD+I + +++ ++ + +
Sbjct: 83 MFHSFKQFPRYVTGACCLFLAGKVEETPKKCKDIIKTARSLLND--------VQFGQFGD 134
Query: 150 QQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQN---ALAQVAWNFVNDGLR 206
KE +++ ER++L T+ FD V+HPY+ L++ K+LK +N L Q+AW FVND L
Sbjct: 135 DPKEEVMVLERILLQTIKFDLQVEHPYQFLLKYAKQLKGDKNKIQKLVQMAWTFVNDSLC 194
Query: 207 TSLCLQFKPQHIAAGAIFLAAKFLNVKLE--------KLWWQVFDVTLTAHQLEEVSNQM 258
T+L LQ++P+ IA ++LA + +++ + WW+ F + LE++ +Q+
Sbjct: 195 TTLSLQWEPEIIAVAVMYLAGRLCKFEIQEWTSKPMYRRWWEQFVQDVPVDVLEDICHQI 254
Query: 259 LELYEQNQ 266
L+LY Q +
Sbjct: 255 LDLYSQGK 262
>CYCK_HUMAN (O75909) Cyclin K
Length = 357
Score = 152 bits (383), Expect = 3e-36
Identities = 82/248 (33%), Positives = 145/248 (58%), Gaps = 20/248 (8%)
Query: 30 WYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFF 89
WY +K++ ++PS+ +G+D EA R+ ++ D+G RL + T+A+ II+ HRF+
Sbjct: 24 WYWDKKDLA-HTPSQLEGLDPATEARYRREGARFIFDVGTRLGLHYDTLATGIIYFHRFY 82
Query: 90 LRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYE 149
+ S + R + C+FLAGKVEETP+ KD+I + +++ ++ + +
Sbjct: 83 MFHSFKQFPRYVTGACCLFLAGKVEETPKKCKDIIKTARSLLND--------VQFGQFGD 134
Query: 150 QQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQN---ALAQVAWNFVNDGLR 206
KE +++ ER++L T+ FD V+HPY+ L++ K+LK +N L Q+AW FVND L
Sbjct: 135 DPKEEVMVLERILLQTIKFDLQVEHPYQFLLKYAKQLKGDKNKIQKLVQMAWTFVNDSLC 194
Query: 207 TSLCLQFKPQHIAAGAIFLAAKFLNVKLE--------KLWWQVFDVTLTAHQLEEVSNQM 258
T+L LQ++P+ IA ++LA + +++ + WW+ F + LE++ +Q+
Sbjct: 195 TTLSLQWEPEIIAVAVMYLAGRLCKFEIQEWTSKPMYRRWWEQFVQDVPVDVLEDICHQI 254
Query: 259 LELYEQNQ 266
L+LY Q +
Sbjct: 255 LDLYSQGK 262
>CG1C_SCHPO (O74627) Cyclin pch1 (Pombe cyclin c homolog 1)
Length = 342
Score = 136 bits (343), Expect = 1e-31
Identities = 73/210 (34%), Positives = 122/210 (57%), Gaps = 3/210 (1%)
Query: 25 ENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIF 84
+N S+W +S+ ++ +PS DG+ L +E R C ++ ++G+RLK+PQ +A+A I+
Sbjct: 13 QNTSQWIISKDQLVF-TPSALDGIPLDQEEIQRSKGCNFIINVGLRLKLPQTALATANIY 71
Query: 85 CHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQ 144
HRF+LR S +A +C+FLA KVE++ R L+D+++ ++ K V +Q
Sbjct: 72 FHRFYLRFSLKNYHYYEVAATCIFLATKVEDSVRKLRDIVINCAKVAQKNSNVLVD--EQ 129
Query: 145 KEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDG 204
+ Y + +++IL E V+L L FDF V+HPY ++ IKK + +VAW ++ND
Sbjct: 130 TKEYWRWRDVILYTEEVLLEALCFDFTVEHPYPYVLSFIKKFVADDKNVTKVAWTYINDS 189
Query: 205 LRTSLCLQFKPQHIAAGAIFLAAKFLNVKL 234
R+ CL + P+ IAA A A + + L
Sbjct: 190 TRSIACLLYSPKTIAAAAFQFALEKNEINL 219
>CCT_DROME (O96433) Cyclin T
Length = 1097
Score = 123 bits (309), Expect = 1e-27
Identities = 80/278 (28%), Positives = 145/278 (51%), Gaps = 20/278 (7%)
Query: 25 ENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIF 84
E WY S ++ NSPS++ G+ E R+ +Q++G RL+V Q+ I +AI++
Sbjct: 40 EKDKIWYFSNDQLA-NSPSRRCGIKGDDELQYRQMTAYLIQEMGQRLQVSQLCINTAIVY 98
Query: 85 CHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQ 144
HRF+ S R +A++ +FLA KVEE PR L+ VI + + + P Q
Sbjct: 99 MHRFYAFHSFTHFHRNSMASASLFLAAKVEEQPRKLEHVIRAANKCL---PPTTEQN--- 152
Query: 145 KEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDG 204
Y + + ++ E V+L TLGFD + HP+ +V + +K ++ LAQ ++ ++
Sbjct: 153 ---YAELAQELVFNENVLLQTLGFDVAIDHPHTHVVRTCQLVKACKD-LAQTSYFLASNS 208
Query: 205 LR-TSLCLQFKPQHIAAGAIFLAAKFLNVKLE-----KLWWQVFDVTLTAHQLEEVSNQM 258
L TS+CLQ++P +A I+LA K+ ++ K W+ D T++ L++++++
Sbjct: 209 LHLTSMCLQYRPTVVACFCIYLACKWSRWEIPQSTEGKHWFYYVDKTVSLDLLKQLTDEF 268
Query: 259 LELYEQNQI---APSNDVEGTAGGGNRATPKASTSNDE 293
+ +YE++ + N ++ A G + T + E
Sbjct: 269 IAIYEKSPARLKSKLNSIKAIAQGASNRTANSKDKPKE 306
>YL34_CAEEL (P34425) Hypothetical protein F44B9.4 in chromosome III
Length = 468
Score = 92.0 bits (227), Expect = 4e-18
Identities = 67/259 (25%), Positives = 128/259 (48%), Gaps = 28/259 (10%)
Query: 28 SRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDL--------GMRLKVPQVTIA 79
++W +++E+++ + S Q+G+ ++E R+ ++Q++ ++K+ +
Sbjct: 18 TKWLFTKEEMKKTA-SIQEGMSREEELASRQMAAAFIQEMIDGLNNVKDPKMKIGHTGLC 76
Query: 80 SAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAV 139
A HRF+ S K D R + +C+FLAGK +E PR L VI V E +KD
Sbjct: 77 VAHTHMHRFYYLHSFKKYDYRDVGAACVFLAGKSQECPRKLSHVISVWRE---RKDR--- 130
Query: 140 QRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKL--KVSQNALAQVA 197
+++ + + ++I+L E ++L T+ FD +V P+ +++ +KK+ K L A
Sbjct: 131 KQLTTETARNEAAQIIVLLESMILQTIAFDLNVHLPHIYVLDIMKKVDKKEHYRPLTSCA 190
Query: 198 WNFVNDGLR-TSLCLQFKPQHIAAGAIFLAAKFLNVKLEKL----------WWQVFDVTL 246
+ F D + T L++ ++ I L A + NV++E+L W+ FD T+
Sbjct: 191 YYFATDVIAVTDWSLRYSAASMSIVIIHLMAAYANVRIERLFADFINEDSPWYAKFDETM 250
Query: 247 TAHQLEEVSNQMLELYEQN 265
T +L E+ L Y +
Sbjct: 251 TNEKLREMEVDFLVTYRNS 269
>YL33_CAEEL (P34424) Hypothetical protein F44B9.3 in chromosome III
Length = 555
Score = 88.6 bits (218), Expect = 5e-17
Identities = 63/267 (23%), Positives = 139/267 (51%), Gaps = 30/267 (11%)
Query: 23 SQENGSR-------WYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRL---K 72
+++NGSR W ++++ ++PS+++G+ ++E R+ ++ D+ M+L K
Sbjct: 9 AEKNGSRYGRMIHPWLRKKQDMLADTPSRREGMTYEEELSKRQQGGVFIFDIAMQLTHGK 68
Query: 73 VPQVTIASAIIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVI--LVSYEI 130
A +RFF S + D R +A +C+FLAGK E+ P+ LK V+ L ++
Sbjct: 69 GEHGLSGVAATLFNRFFNVHSLKRCDFRDVAAACVFLAGKNEDAPKKLKYVVTQLWQFKY 128
Query: 131 IHKKDPAAVQRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQ 190
H K + + ++ + Q ++ L E V+L T+ FD +V P++ +++ ++ ++ +
Sbjct: 129 PHNK------QFQSEQHFLDQCNVVTLIEDVLLKTISFDINVDLPHQYVLKLMRDVEKGR 182
Query: 191 NA---LAQVAWNFVNDGL-RTSLCLQFKPQHIAAGAIFLAAKFLNVKLEKL--------W 238
N + + A+ D L T +++ IA + +AA F N+ ++ + W
Sbjct: 183 NVYKDMVKTAYYMATDVLIITDWSVRYSCASIATACVNIAAFFHNINMDDIVPFELSDRW 242
Query: 239 WQVFDVTLTAHQLEEVSNQMLELYEQN 265
+++ D ++T ++E ++ + L+++ +N
Sbjct: 243 YRLEDQSMTREEVEAMTKEFLDIFSRN 269
>CG1C_ORYSA (P93411) G1/S-specific cyclin C-type
Length = 257
Score = 85.5 bits (210), Expect = 4e-16
Identities = 64/239 (26%), Positives = 122/239 (50%), Gaps = 17/239 (7%)
Query: 32 LSRKEIEENSPSKQD-GVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFL 90
L ++++++ + D G+ L++ ++ ++ L ++KV Q IA+A+ + R +
Sbjct: 15 LDQEDVDKVPQADSDRGITLEEFRLVKIHMSFHIWRLAQQVKVRQRVIATAVTYFRRVYT 74
Query: 91 RQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYEQ 150
R+S + D R++A +C++LA KVEE+ ++ +LV Y + E Y
Sbjct: 75 RKSMTEYDPRLVAPTCLYLASKVEES--TVQARLLVFY---------IKKMCASDEKYRF 123
Query: 151 QKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTSLC 210
+ + IL E +L L + V HPY+PL++ ++ ++ L Q AW VND + L
Sbjct: 124 EIKDILEMEMKLLEALDYYLVVYHPYRPLLQLLQDAGITD--LTQFAWGIVNDTYKMDLI 181
Query: 211 LQFKPQHIAAGAIFLAAKFLNVKLEKLWWQVFDVTLTAHQLEEVSNQMLELYEQNQIAP 269
L P IA I++A+ L K LW++ V + ++ +S ++L+ Y+ +I P
Sbjct: 182 LIHPPYMIALACIYIAS-VLKDKDITLWFEELRVDMNI--VKNISMEILDFYDTYKIDP 237
>CG1C_CHICK (P55168) Cyclin C
Length = 283
Score = 85.5 bits (210), Expect = 4e-16
Identities = 71/244 (29%), Positives = 123/244 (50%), Gaps = 24/244 (9%)
Query: 29 RWYLSRKEIEENSPSKQDGVDLKKEAY--LRKSYCTYLQDLGMRLKVPQVTIASAIIFCH 86
+W L ++++ + ++D L +E Y L+ + +Q LG LK+ Q IA+A ++
Sbjct: 13 QWILDKQDLLKER--QKDLKFLSEEEYWKLQIFFTNVIQALGEHLKLRQQVIATATVYFK 70
Query: 87 RFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKE 146
RF+ R S D ++A +C+FLA KVEE + + L+S AA +K +
Sbjct: 71 RFYARYSLKSIDPVLMAPTCVFLASKVEEF-GVVSNTRLIS---------AATSVLKTRF 120
Query: 147 VYEQQKEL------ILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNF 200
Y KE IL E +L + V HPY+PL++ ++ + ++ L +AW
Sbjct: 121 SYAFPKEFPYRMNHILECEFYLLELMDCCLIVYHPYRPLLQYVQDMG-QEDMLLPLAWRI 179
Query: 201 VNDGLRTSLCLQFKPQHIAAGAIFLAAKFLNVKLEKLWWQVFDVTLTAHQLEEVSNQMLE 260
VND RT LCL + P IA + +A + K + W+ ++++ ++ E+ +L+
Sbjct: 180 VNDTYRTDLCLLYPPFMIALACLHVAC-VVQQKDARQWFA--ELSVDMEKILEIIRVILK 236
Query: 261 LYEQ 264
LYEQ
Sbjct: 237 LYEQ 240
>CG1C_HUMAN (P24863) Cyclin C
Length = 303
Score = 84.7 bits (208), Expect = 7e-16
Identities = 70/244 (28%), Positives = 123/244 (49%), Gaps = 24/244 (9%)
Query: 29 RWYLSRKEIEENSPSKQDGVDLKKEAY--LRKSYCTYLQDLGMRLKVPQVTIASAIIFCH 86
+W L ++++ + ++D L +E Y L+ + +Q LG LK+ Q IA+A ++
Sbjct: 33 QWILDKQDLLKER--QKDLKFLSEEEYWKLQIFFTNVIQALGEHLKLRQQVIATATVYFK 90
Query: 87 RFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKE 146
RF+ R S D ++A +C+FLA KVEE ++ + +I AA +K +
Sbjct: 91 RFYARYSLKSIDPVLMAPTCVFLASKVEEFG------VVSNTRLI----AAATSVLKTRF 140
Query: 147 VYEQQKEL------ILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNF 200
Y KE IL E +L + V HPY+PL++ ++ + ++ L +AW
Sbjct: 141 SYAFPKEFPYRMNHILECEFYLLELMDCCLIVYHPYRPLLQYVQDMG-QEDMLLPLAWRI 199
Query: 201 VNDGLRTSLCLQFKPQHIAAGAIFLAAKFLNVKLEKLWWQVFDVTLTAHQLEEVSNQMLE 260
VND RT LCL + P IA + +A + K + W+ ++++ ++ E+ +L+
Sbjct: 200 VNDTYRTDLCLLYPPFMIALACLHVAC-VVQQKDARQWFA--ELSVDMEKILEIIRVILK 256
Query: 261 LYEQ 264
LYEQ
Sbjct: 257 LYEQ 260
>CG1C_DROME (P25008) G1/S-specific cyclin C
Length = 267
Score = 84.7 bits (208), Expect = 7e-16
Identities = 66/248 (26%), Positives = 116/248 (46%), Gaps = 27/248 (10%)
Query: 26 NGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKS--YCTYLQDLGMRLKVPQVTIASAII 83
+ +W L + ++ + D + L ++ Y + + +Q LG +LK+ Q IA+A +
Sbjct: 10 HSQQWILDKPDLLRER--QHDLLALNEDEYQKVFIFFANVIQVLGEQLKLRQQVIATATV 67
Query: 84 FCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAV--QR 141
+ RF+ R S D ++A +C+ LA KVEE + +I ++
Sbjct: 68 YFKRFYARNSLKNIDPLLLAPTCILLASKVEE------------FGVISNSRLISICQSA 115
Query: 142 IKQKEVYEQQKEL------ILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQ 195
IK K Y +E IL E +L L V PY+PL++ ++ + ++ L
Sbjct: 116 IKTKFSYAYAQEFPYRTNHILECEFYLLENLDCCLIVYQPYRPLLQLVQDMG-QEDQLLT 174
Query: 196 VAWNFVNDGLRTSLCLQFKPQHIAAGAIFLAAKFLNVKLEKLWWQVFDVTLTAHQLEEVS 255
++W VND LRT +CL + P IA + +A L K W+ +V L +++E+
Sbjct: 175 LSWRIVNDSLRTDVCLLYPPYQIAIACLQIACVILQKDATKQWFAELNVDL--DKVQEIV 232
Query: 256 NQMLELYE 263
++ LYE
Sbjct: 233 RAIVNLYE 240
>CG1C_RAT (P39947) Cyclin C
Length = 298
Score = 82.8 bits (203), Expect = 2e-15
Identities = 66/223 (29%), Positives = 111/223 (49%), Gaps = 22/223 (9%)
Query: 50 LKKEAY--LRKSYCTYLQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKNDRRIIATSCM 107
L +E Y L+ + +Q LG LK+ Q IA+A ++ RF+ R S D ++A +C+
Sbjct: 47 LSEEEYWKLQIFFTNVIQALGEHLKLRQQVIATATVYFKRFYARYSLKSIDPVLMAPTCV 106
Query: 108 FLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKEVYEQQKEL------ILLGERV 161
FLA KVEE ++ + +I A +K + Y KE IL E
Sbjct: 107 FLASKVEEFG------VVSNTSLI----AATTSVLKTRFSYASPKEFPYRMNHILECEFY 156
Query: 162 VLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNFVNDGLRTSLCLQFKPQHIAAG 221
+L + V HPY+PL++ ++ + ++ L +AW VND RT LCL + P IA
Sbjct: 157 LLELMDCCLIVYHPYRPLLQYVQDMG-QEDVLLPLAWRIVNDTYRTDLCLLYPPFMIALA 215
Query: 222 AIFLAAKFLNVKLEKLWWQVFDVTLTAHQLEEVSNQMLELYEQ 264
+ +A + K + W+ ++++ ++ E+ +L+LYEQ
Sbjct: 216 CLHVAC-VVQQKDARQWFA--ELSVDMEKILEIIRVILKLYEQ 255
>CG1C_MOUSE (Q62447) Cyclin C
Length = 304
Score = 81.3 bits (199), Expect = 7e-15
Identities = 69/244 (28%), Positives = 121/244 (49%), Gaps = 24/244 (9%)
Query: 29 RWYLSRKEIEENSPSKQDGVDLKKEAY--LRKSYCTYLQDLGMRLKVPQVTIASAIIFCH 86
+W L ++++ + ++D L +E Y L+ + +Q LG LK+ Q IA+A ++
Sbjct: 34 QWILDKQDLLKER--QKDLKFLSEEEYWKLQIFFTNVIQALGEHLKLRQQVIATATVYFK 91
Query: 87 RFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKE 146
RF R S D ++A +C+FLA KVEE ++ + +I A +K +
Sbjct: 92 RFDARYSLKSIDPVLMAPTCVFLASKVEEFG------VVSNTRLI----AATTSVLKTRF 141
Query: 147 VYEQQKEL------ILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQNALAQVAWNF 200
Y KE IL E +L + V HPY+PL++ ++ + ++ L +AW
Sbjct: 142 SYAFPKEFPYRMNHILECEFYLLELMDCCLIVYHPYRPLLQYVQDMG-QEDVLLPLAWRI 200
Query: 201 VNDGLRTSLCLQFKPQHIAAGAIFLAAKFLNVKLEKLWWQVFDVTLTAHQLEEVSNQMLE 260
VND RT LCL + P IA + +A + K + W+ ++++ ++ E+ +L+
Sbjct: 201 VNDTYRTDLCLLYPPFMIALACLHVAC-VVQQKDARQWFA--ELSVDMEKILEIIRVILK 257
Query: 261 LYEQ 264
LYEQ
Sbjct: 258 LYEQ 261
>UME3_YEAST (P47821) RNA polymerase II holoenzyme cyclin-like
subunit
Length = 323
Score = 65.5 bits (158), Expect = 4e-10
Identities = 55/211 (26%), Positives = 99/211 (46%), Gaps = 28/211 (13%)
Query: 23 SQENGSRWYLSRKEIEENSPSKQDGVDLKKEAYLRKSYCTYL-QDLGMRLKVPQVTIASA 81
S++NG + I +N P + K+ LR YC +L LG RL + Q +A+A
Sbjct: 46 SKQNGIE-----QSITKNIPITHRDLHYDKDYNLR-IYCYFLIMKLGRRLNIRQYALATA 99
Query: 82 IIFCHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQR 141
I+ RF ++ S + + ++ T+C++LA KVEE P+ ++ ++ + + + P +
Sbjct: 100 HIYLSRFLIKASVREINLYMLVTTCVYLACKVEECPQYIRTLVSEARTLWPEFIPPDPTK 159
Query: 142 IKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLK-------VSQNALA 194
+ + E Y +L L V HPY+ L + ++ LK +S + L
Sbjct: 160 VTEFEFY-------------LLEELESYLIVHHPYQSLKQIVQVLKQPPFQITLSSDDL- 205
Query: 195 QVAWNFVNDGLRTSLCLQFKPQHIAAGAIFL 225
Q W+ +ND + L + P IA +F+
Sbjct: 206 QNCWSLINDSYINDVHLLYPPHIIAVACLFI 236
>SRBB_SCHPO (O94503) RNA polymerase II holoenzyme cyclin-like
subunit (Suppressor of RNA polymerase B srb11)
Length = 228
Score = 63.9 bits (154), Expect = 1e-09
Identities = 42/164 (25%), Positives = 83/164 (50%), Gaps = 15/164 (9%)
Query: 64 LQDLGMRLKVPQVTIASAIIFCHRFFLRQSHAKN-DRRIIATSCMFLAGKVEETPRPLKD 122
+Q G RL++ Q +A+AI+ R+ L+++ K + +C++L+ KVEE P ++
Sbjct: 39 VQTFGDRLRLRQRVLATAIVLLRRYMLKKNEEKGFSLEALVATCIYLSCKVEECPVHIRT 98
Query: 123 VILVSYEIIHKKDPAAVQRIKQKEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEA 182
+ + ++ K ++ + + E + E+I + + ++ V HPY L +A
Sbjct: 99 ICNEANDLWSLK-----VKLSRSNISEIEFEIISVLDAFLI--------VHHPYTSLEQA 145
Query: 183 IKKLKVSQNALAQVAWNFVNDGLRTSLCLQFKPQHIAAGAIFLA 226
++Q L + AW+ VND +SLCL P +A A+ ++
Sbjct: 146 FHDGIINQKQL-EFAWSIVNDSYASSLCLMAHPHQLAYAALLIS 188
>CTK2_YEAST (P46962) CTD kinase beta subunit (CTD kinase 38 kDa
subunit) (CTDK-I beta subunit)
Length = 323
Score = 54.7 bits (130), Expect = 7e-07
Identities = 56/206 (27%), Positives = 95/206 (45%), Gaps = 20/206 (9%)
Query: 28 SRWYLSRKEIE---ENSPSKQDGVDLKKEAYLRKSYCTYLQDLGMRLKVPQVTIASAIIF 84
SR +LS+++I+ +N+ S + KK A + +L DL ++LK P+ T+ +A+ F
Sbjct: 12 SRPFLSKRQIQRAQKNTISDYRNYNQKKLAVFK-----FLSDLCVQLKFPRKTLETAVYF 66
Query: 85 CHRFFLRQSHAKNDRRIIATSCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQ 144
R+ L +ATSC+ L K ET + D+ +S + + V +I
Sbjct: 67 YQRYHLFNRFETEVCYTVATSCLTLGCKEVETIKKTNDICTLSLRLRN------VVKI-N 119
Query: 145 KEVYEQQKELILLGERVVLATLGFDFHVQHPYKPLVEAIKKLKVSQN---ALAQVAWNFV 201
++ E K+ + E +L + FD+ V + Y + E + K+ + L +AW
Sbjct: 120 TDILENFKKRVFQIELRILESCSFDYRVNN-YVHIDEYVIKIGRELSFDYKLCNLAWVIA 178
Query: 202 NDGLRTSLCLQFKPQHIAAGAIFLAA 227
D L+ L PQH A AI A
Sbjct: 179 YDALKLETILVI-PQHSIALAILKIA 203
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.130 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,291,301
Number of Sequences: 164201
Number of extensions: 3285415
Number of successful extensions: 10028
Number of sequences better than 10.0: 401
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 355
Number of HSP's that attempted gapping in prelim test: 9592
Number of HSP's gapped (non-prelim): 690
length of query: 629
length of database: 59,974,054
effective HSP length: 116
effective length of query: 513
effective length of database: 40,926,738
effective search space: 20995416594
effective search space used: 20995416594
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0176a.15


