

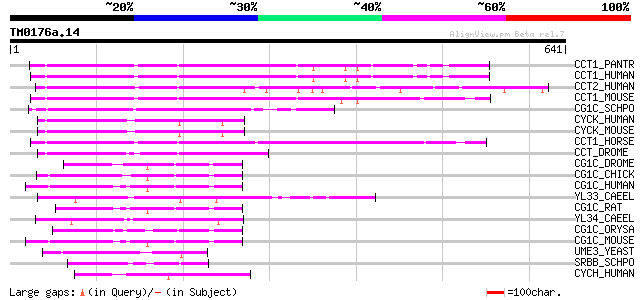
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.14
(641 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CCT1_PANTR (Q8HXN7) Cyclin T1 (Cyclin T) (CycT1) 161 4e-39
CCT1_HUMAN (O60563) Cyclin T1 (Cyclin T) (CycT1) 161 4e-39
CCT2_HUMAN (O60583) Cyclin T2 160 1e-38
CCT1_MOUSE (Q9QWV9) Cyclin T1 (Cyclin T) (CycT1) 157 6e-38
CG1C_SCHPO (O74627) Cyclin pch1 (Pombe cyclin c homolog 1) 155 2e-37
CYCK_HUMAN (O75909) Cyclin K 155 3e-37
CYCK_MOUSE (O88874) Cyclin K 153 1e-36
CCT1_HORSE (Q9XT26) Cyclin T1 (Cyclin T) (CycT1) 153 2e-36
CCT_DROME (O96433) Cyclin T 125 3e-28
CG1C_DROME (P25008) G1/S-specific cyclin C 91 7e-18
CG1C_CHICK (P55168) Cyclin C 91 9e-18
CG1C_HUMAN (P24863) Cyclin C 91 1e-17
YL33_CAEEL (P34424) Hypothetical protein F44B9.3 in chromosome III 90 2e-17
CG1C_RAT (P39947) Cyclin C 90 2e-17
YL34_CAEEL (P34425) Hypothetical protein F44B9.4 in chromosome III 89 3e-17
CG1C_ORYSA (P93411) G1/S-specific cyclin C-type 88 6e-17
CG1C_MOUSE (Q62447) Cyclin C 87 1e-16
UME3_YEAST (P47821) RNA polymerase II holoenzyme cyclin-like sub... 72 4e-12
SRBB_SCHPO (O94503) RNA polymerase II holoenzyme cyclin-like sub... 72 6e-12
CYCH_HUMAN (P51946) Cyclin H (MO15-associated protein) (p37) (p34) 60 1e-08
>CCT1_PANTR (Q8HXN7) Cyclin T1 (Cyclin T) (CycT1)
Length = 725
Score = 161 bits (408), Expect = 4e-39
Identities = 148/550 (26%), Positives = 244/550 (43%), Gaps = 37/550 (6%)
Query: 24 QDNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIAT 83
+ ++ RWYF+R+++E NSPS++ GVD KE R+ LQD+G RL V Q+TI T
Sbjct: 2 EGERKNNKRWYFTREQLE-NSPSRRFGVDPDKELSYRQQAANLLQDMGQRLNVSQLTINT 60
Query: 84 AIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQ 143
AI++ HRF++ QS + +A +FLA KVEE P+ L+ VI V++ +H ++
Sbjct: 61 AIVYMHRFYMIQSFTQFPGNSVAPAALFLAAKVEEQPKKLEHVIKVAHTCLHPQESLPDT 120
Query: 144 RIKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNF 203
R + Y QQ + +++ E ++L TLGF+L + HP+ +V+ + + +++ LAQ ++
Sbjct: 121 R---SEAYLQQVQDLVILESIILQTLGFELTIDHPHTHVVKCTQLVRASKD-LAQTSYFM 176
Query: 204 VNDGLR-TSLCLQFKPHHIAAGAIFLAAKFLKVKLP-SDGEKVWWQEFD--VTPRQLEEV 259
+ L T+ LQ+ P +A I LA K+ ++P S K WW+ D VT L+E+
Sbjct: 177 ATNSLHLTTFSLQYTPPVVACVCIHLACKWSNWEIPVSTDGKHWWEYVDATVTLELLDEL 236
Query: 260 SNQMLELYEQNRMAQSNDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETSK 319
+++ L++ E+ + T DE S L++ SS+ T
Sbjct: 237 THEFLQILEKTPNRLKRIWNWRACEAAKKTKADDRGTDEKTSEQTILNMISQSSSDTTIA 296
Query: 320 PATSKNVFVSSANHVGRPVSNHGVSGSTEV-----KHPVEDDVKGNLHPKQEPSAYEENM 374
S + +SA PVS S T V K + L P Q E
Sbjct: 297 GLMSMSTSTTSA-VPSLPVSEESSSNLTSVEMLPGKRWLSSQPSFKLEPTQGHRTSENLA 355
Query: 375 QEAQDMVRSRSG---YAKGNQHPKQEPST----YEEKMQEAQDMVRSRSGYGKEQESNVG 427
D + G + Q+ K PS E + + A+++ + + E+NV
Sbjct: 356 LTGVDHSLPQDGSNAFISQKQNSKSVPSAKVSLKEYRAKHAEELAAQKRQL-ENMEANVK 414
Query: 428 R--SNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNIS 485
+ + + +D HSS L +P ++K D K AL+ R G
Sbjct: 415 SQYAYAAQNLLSHHDSHSSVILKMPIEGSENPERPFLEKAD----KTALKMRIPVAG--G 468
Query: 486 KKTELVDDEDLIERVRELEDGIELAAQSEKNKQDG-RKSLSKSLDRSAFENMQHGKHQDH 544
K E++ R++ + AA + +D KS + H H H
Sbjct: 469 DKAASSKPEEIKMRIK-----VHAAADKHNSVEDSVTKSREHKEKHKTHPSNHHHHHNHH 523
Query: 545 SEEHLHLVKP 554
S +H H P
Sbjct: 524 SHKHSHSQLP 533
>CCT1_HUMAN (O60563) Cyclin T1 (Cyclin T) (CycT1)
Length = 726
Score = 161 bits (408), Expect = 4e-39
Identities = 148/549 (26%), Positives = 243/549 (43%), Gaps = 37/549 (6%)
Query: 25 DNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATA 84
+ + RWYF+R+++E NSPS++ GVD KE R+ LQD+G RL V Q+TI TA
Sbjct: 4 ERKNNNKRWYFTREQLE-NSPSRRFGVDPDKELSYRQQAANLLQDMGQRLNVSQLTINTA 62
Query: 85 IIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQR 144
I++ HRF++ QS + +A +FLA KVEE P+ L+ VI V++ +H ++ R
Sbjct: 63 IVYMHRFYMIQSFTQFPGNSVAPAALFLAAKVEEQPKKLEHVIKVAHTCLHPQESLPDTR 122
Query: 145 IKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFV 204
+ Y QQ + +++ E ++L TLGF+L + HP+ +V+ + + +++ LAQ ++
Sbjct: 123 ---SEAYLQQVQDLVILESIILQTLGFELTIDHPHTHVVKCTQLVRASKD-LAQTSYFMA 178
Query: 205 NDGLR-TSLCLQFKPHHIAAGAIFLAAKFLKVKLP-SDGEKVWWQEFD--VTPRQLEEVS 260
+ L T+ LQ+ P +A I LA K+ ++P S K WW+ D VT L+E++
Sbjct: 179 TNSLHLTTFSLQYTPPVVACVCIHLACKWSNWEIPVSTDGKHWWEYVDATVTLELLDELT 238
Query: 261 NQMLELYEQNRMAQSNDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETSKP 320
++ L++ E+ + T DE S L++ SS+ T
Sbjct: 239 HEFLQILEKTPNRLKRIWNWRACEAAKKTKADDRGTDEKTSEQTILNMISQSSSDTTIAG 298
Query: 321 ATSKNVFVSSANHVGRPVSNHGVSGSTEV-----KHPVEDDVKGNLHPKQEPSAYEENMQ 375
S + +SA PVS S T V K + L P Q E
Sbjct: 299 LMSMSTSTTSA-VPSLPVSEESSSNLTSVEMLPGKRWLSSQPSFKLEPTQGHRTSENLAL 357
Query: 376 EAQDMVRSRSG---YAKGNQHPKQEPST----YEEKMQEAQDMVRSRSGYGKEQESNVGR 428
D + G + Q+ K PS E + + A+++ + + E+NV
Sbjct: 358 TGVDHSLPQDGSNAFISQKQNSKSVPSAKVSLKEYRAKHAEELAAQKRQL-ENMEANVKS 416
Query: 429 --SNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNISK 486
+ + + +D HSS L +P ++K D K AL+ R G
Sbjct: 417 QYAYAAQNLLSHHDSHSSVILKMPIEGSENPERPFLEKAD----KTALKMRIPVAG--GD 470
Query: 487 KTELVDDEDLIERVRELEDGIELAAQSEKNKQDG-RKSLSKSLDRSAFENMQHGKHQDHS 545
K E++ R++ + AA + +D KS + H H HS
Sbjct: 471 KAASSKPEEIKMRIK-----VHAAADKHNSVEDSVTKSREHKEKHKTHPSNHHHHHNHHS 525
Query: 546 EEHLHLVKP 554
+H H P
Sbjct: 526 HKHSHSQLP 534
>CCT2_HUMAN (O60583) Cyclin T2
Length = 730
Score = 160 bits (405), Expect = 1e-38
Identities = 164/670 (24%), Positives = 291/670 (42%), Gaps = 95/670 (14%)
Query: 31 SRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHR 90
SRW+F+R+++E N+PS++ GV+ KE R+ +Q++G RL V Q+TI TAI++ HR
Sbjct: 9 SRWFFTREQLE-NTPSRRCGVEADKELSCRQQAANLIQEMGQRLNVSQLTINTAIVYMHR 67
Query: 91 FFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDV 150
F++ S K ++ II++ +FLA KVEE R L+ VI V++ +H +P + D
Sbjct: 68 FYMHHSFTKFNKNIISSTALFLAAKVEEQARKLEHVIKVAHACLHPLEPLLDTKC---DA 124
Query: 151 YEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLR- 209
Y QQ + +++ E ++L TLGF++ + HP+ +V+ + + +++ LAQ ++ + L
Sbjct: 125 YLQQTQELVILETIMLQTLGFEITIEHPHTDVVKCTQLVRASKD-LAQTSYFMATNSLHL 183
Query: 210 TSLCLQFKPHHIAAGAIFLAAKFLKVKLP-SDGEKVWWQEFD--VTPRQLEEVSNQMLEL 266
T+ CLQ+KP IA I LA K+ ++P S K WW+ D VT L+E++++ L++
Sbjct: 184 TTFCLQYKPTVIACVCIHLACKWSNWEIPVSTDGKHWWEYVDPTVTLELLDELTHEFLQI 243
Query: 267 YEQ-------------NRMAQSNDVEGTTGGGNRATPKAPTS-------NDEAASTNRNL 306
E+ N+ A+ V+G TP +S D N
Sbjct: 244 LEKTPNRLKKIRNWRANQAARKPKVDGQVS----ETPLLGSSLVQNSILVDSVTGVPTNP 299
Query: 307 HIGGPS-STLETSKPATSKNVFVSSAN--------HVGRPVSNHGVSGSTE--------- 348
PS S P S N+ V ++ G P +++G+S E
Sbjct: 300 SFQKPSTSAFPAPVPLNSGNISVQDSHTSDNLSMLATGMPSTSYGLSSHQEWPQHQDSAR 359
Query: 349 ----VKHPVEDDVKG---NLHPKQEPS-AYEENMQEAQDMVRSRSGYAKGNQHPKQEPST 400
E + G N++ +Q PS + + D + S + H K S
Sbjct: 360 TEQLYSQKQETSLSGSQYNINFQQGPSISLHSGLHHRPDKISDHSSVKQEYTH-KAGSSK 418
Query: 401 YEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDVELNDKHSSKNLNHR------DATF 454
+ + ++ + K +E + ++ D+++ D + + + + A
Sbjct: 419 HHGPISTTPGIIPQKMSLDKYRE----KRKLETLDLDVRDHYIAAQVEQQHKQGQSQAAS 474
Query: 455 ASPNLEGIK-KIDRDKVKAALEKRKKATGNISKKTELVDDEDLIERVRELEDGIELAAQS 513
+S IK KI + + +K+ +G++ + + D EL+ I++++
Sbjct: 475 SSSVTSPIKMKIPIANTEKYMADKKEKSGSLKLRIP-IPPTDKSASKEELKMKIKVSSSE 533
Query: 514 EKNKQDGRKSLSKSLDRSAFENMQH-GKHQDH--SEEHLHLVKPSHEADLSAVEEGEVSA 570
+ D + KS S + H KH++H S H K SH S+ + SA
Sbjct: 534 RHSSSD--EGSGKSKHSSPHISRDHKEKHKEHPSSRHHTSSHKHSHSHSGSSSGGSKHSA 591
Query: 571 ---------------LDDIDLGPKSSNQKRKVESSPDNFVEGKKRHNYGSGSTHHNRI-- 613
D I SS ++ V + N + + SGS+ +
Sbjct: 592 DGIPPTVLRSPVGLSSDGISSSSSSSRKRLHVNDASHNHHSKMSKSSKSSGSSSSSSSSV 651
Query: 614 -DYIEDRNKV 622
YI N V
Sbjct: 652 KQYISSHNSV 661
>CCT1_MOUSE (Q9QWV9) Cyclin T1 (Cyclin T) (CycT1)
Length = 724
Score = 157 bits (398), Expect = 6e-38
Identities = 142/550 (25%), Positives = 246/550 (43%), Gaps = 38/550 (6%)
Query: 25 DNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATA 84
+ + RWYF+R+++E NSPS++ GVD KE R+ LQD+G RL V Q+TI TA
Sbjct: 4 ERKNNNKRWYFTREQLE-NSPSRRFGVDSDKELSYRQQAANLLQDVGQRLNVSQLTINTA 62
Query: 85 IIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQR 144
I++ HRF++ QS + R +A +FLA KVEE P+ L+ VI V++ +H ++ R
Sbjct: 63 IVYMHRFYMIQSFTQFHRYSMAPAALFLAAKVEEQPKKLEHVIKVAHTCLHPQESLPDTR 122
Query: 145 IKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFV 204
+ Y QQ + +++ E ++L TLGF+L + HP+ +V+ + + +++ LAQ ++
Sbjct: 123 ---SEAYLQQVQDLVILESIILQTLGFELTIDHPHTHVVKCTQLVRASKD-LAQTSYFMA 178
Query: 205 NDGLR-TSLCLQFKPHHIAAGAIFLAAKFLKVKLP-SDGEKVWWQEFD--VTPRQLEEVS 260
+ L T+ LQ+ P +A I LA K+ ++P S K WW+ D VT L+E++
Sbjct: 179 TNSLHLTTFSLQYTPPVVACVCIHLACKWSNWEIPVSTDGKHWWEYVDATVTLELLDELT 238
Query: 261 NQMLELYEQNRMAQSNDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETSKP 320
++ L++ E+ T DE S L++ +S+ T
Sbjct: 239 HEFLQILEKTPSRLKRIRNWRAYQAAMKTKPDDRGADENTSEQTILNMISQTSSDTTIAG 298
Query: 321 ATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQEAQDM 380
S + +SA P S S T V + + P + +A E+ +
Sbjct: 299 LMSMSTASTSA-VPSLPSSEESSSSLTSVDMLQGERWLSSQPPFKLEAAQGHRTSESLAL 357
Query: 381 V--------RSRSGYAKGNQHPKQEPST----YEEKMQEAQDMVRSRSGYGKEQESNVGR 428
+ S + Q K PS E + + A+++ + + + +
Sbjct: 358 IGVDHSLQQDGSSAFGSQKQASKSVPSAKVSLKEYRAKHAEELAAQKRQLENMEANVKSQ 417
Query: 429 SNIKEEDVELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGN--ISK 486
+++ +D HSS L + +P + K D+ +K L A+G+ +S
Sbjct: 418 YAYAAQNLLSHDSHSSVILKMPIESSENPERPFLDKADKSALKMRL---PVASGDKAVSS 474
Query: 487 KTELVDDEDLIERVRELEDGIE-LAAQSEKNKQDGRKSLSKSLDRSAFENMQHGKHQDHS 545
K E + + + + IE +S ++K+ R S H H HS
Sbjct: 475 KPEEIKMRIKVHSAGDKHNSIEDSVTKSREHKEKQRTHPSN----------HHHHHNHHS 524
Query: 546 EEHLHLVKPS 555
H HL P+
Sbjct: 525 HRHSHLQLPA 534
>CG1C_SCHPO (O74627) Cyclin pch1 (Pombe cyclin c homolog 1)
Length = 342
Score = 155 bits (393), Expect = 2e-37
Identities = 110/357 (30%), Positives = 177/357 (48%), Gaps = 32/357 (8%)
Query: 22 GSQDNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTI 81
GSQ+ S+W S+ ++ +PS DG+ L +E R C F+ ++G+RLK+PQ +
Sbjct: 11 GSQNT----SQWIISKDQLVF-TPSALDGIPLDQEEIQRSKGCNFIINVGLRLKLPQTAL 65
Query: 82 ATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAA 141
ATA I+ HRF+LR S +A C+FLA KVE++ R L+D+++ ++ K
Sbjct: 66 ATANIYFHRFYLRFSLKNYHYYEVAATCIFLATKVEDSVRKLRDIVINCAKVAQKNSNVL 125
Query: 142 VQRIKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAW 201
V +Q Y + +++IL E V+L L FD V HPY ++ IKKF + +VAW
Sbjct: 126 VD--EQTKEYWRWRDVILYTEEVLLEALCFDFTVEHPYPYVLSFIKKFVADDKNVTKVAW 183
Query: 202 NFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKL--PSDGEKVWWQEFDVTPRQLEEV 259
++ND R+ CL + P IAA A A + ++ L +DG VW +E V+ ++ V
Sbjct: 184 TYINDSTRSIACLLYSPKTIAAAAFQFALEKNEINLSTTTDGLPVWMEESQVSYEDVKGV 243
Query: 260 SNQMLELYEQ-NRMAQSNDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETS 318
+ LY++ N Q+ ++ G+ A+ AP G PSS S
Sbjct: 244 LTLIDSLYKKINPSKQALPID--QKNGSHASSVAP---------------GTPSSLASVS 286
Query: 319 KPATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQ 375
AT ++ N GR S H ++ T K V+D + ++ S ++ M+
Sbjct: 287 TQATPQH-----QNSSGRTDSFHSLNTETPSKSTVDDQILSTAAQPKKSSDTDKEME 338
>CYCK_HUMAN (O75909) Cyclin K
Length = 357
Score = 155 bits (392), Expect = 3e-37
Identities = 88/248 (35%), Positives = 145/248 (57%), Gaps = 18/248 (7%)
Query: 33 WYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFF 92
WY+ +K++ ++PS+ +G+D EA R+ F+ D+G RL + T+AT II+ HRF+
Sbjct: 24 WYWDKKDLA-HTPSQLEGLDPATEARYRREGARFIFDVGTRLGLHYDTLATGIIYFHRFY 82
Query: 93 LRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYE 152
+ S + R + C+FLAGKVEETP+ KD+I + +++ ++ +
Sbjct: 83 MFHSFKQFPRYVTGACCLFLAGKVEETPKKCKDIIKTARSLLND--------VQFGQFGD 134
Query: 153 QQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQN---ALAQVAWNFVNDGLR 209
KE +++ ER++L T+ FDL V HPY+ L++ K+ K +N L Q+AW FVND L
Sbjct: 135 DPKEEVMVLERILLQTIKFDLQVEHPYQFLLKYAKQLKGDKNKIQKLVQMAWTFVNDSLC 194
Query: 210 TSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKV----WWQEF--DVTPRQLEEVSNQM 263
T+L LQ++P IA ++LA + K ++ K WW++F DV LE++ +Q+
Sbjct: 195 TTLSLQWEPEIIAVAVMYLAGRLCKFEIQEWTSKPMYRRWWEQFVQDVPVDVLEDICHQI 254
Query: 264 LELYEQNR 271
L+LY Q +
Sbjct: 255 LDLYSQGK 262
>CYCK_MOUSE (O88874) Cyclin K
Length = 554
Score = 153 bits (387), Expect = 1e-36
Identities = 87/248 (35%), Positives = 144/248 (57%), Gaps = 18/248 (7%)
Query: 33 WYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFF 92
WY+ +K++ ++PS+ +G+D EA R+ F+ D+G RL + T+A II+ HRF+
Sbjct: 24 WYWDKKDLA-HTPSQLEGLDPATEARYRREGARFIFDVGTRLGLHYDTLANGIIYFHRFY 82
Query: 93 LRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYE 152
+ S + R + C+FLAGKVEETP+ KD+I + +++ ++ +
Sbjct: 83 MFHSFKQFPRYVTGACCLFLAGKVEETPKKCKDIIKTARSLLND--------VQFGQFGD 134
Query: 153 QQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQN---ALAQVAWNFVNDGLR 209
KE +++ ER++L T+ FDL V HPY+ L++ K+ K +N L Q+AW FVND L
Sbjct: 135 DPKEEVMVLERILLQTIKFDLQVEHPYQFLLKYAKQLKGDKNKIQKLVQMAWTFVNDSLC 194
Query: 210 TSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKV----WWQEF--DVTPRQLEEVSNQM 263
T+L LQ++P IA ++LA + K ++ K WW++F DV LE++ +Q+
Sbjct: 195 TTLSLQWEPEIIAVAVMYLAGRLCKFEIQEWTSKPMYRRWWEQFVQDVPVDVLEDICHQI 254
Query: 264 LELYEQNR 271
L+LY Q +
Sbjct: 255 LDLYSQGK 262
>CCT1_HORSE (Q9XT26) Cyclin T1 (Cyclin T) (CycT1)
Length = 727
Score = 153 bits (386), Expect = 2e-36
Identities = 136/535 (25%), Positives = 252/535 (46%), Gaps = 26/535 (4%)
Query: 25 DNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATA 84
+ + RWYF+R+++E NSPS++ G+D KE R+ LQD+G RL V Q+TI TA
Sbjct: 4 ERKNNNKRWYFTREQLE-NSPSRRFGLDPDKELSYRQQAANLLQDMGQRLNVSQLTINTA 62
Query: 85 IIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQR 144
I++ HRF++ QS + R +A +FLA KVEE P+ L+ VI V++ +H ++ R
Sbjct: 63 IVYMHRFYMIQSFTQFHRNSVAPAALFLAAKVEEQPKKLEHVIKVAHACLHPQESLPDTR 122
Query: 145 IKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFV 204
+ Y QQ + +++ E ++L TLGF+L + HP+ +V+ + + +++ LAQ ++
Sbjct: 123 ---SEAYLQQVQDLVILESIILQTLGFELTIDHPHTHVVKCTQLVRASKD-LAQTSYFMA 178
Query: 205 NDGLR-TSLCLQFKPHHIAAGAIFLAAKFLKVKLP-SDGEKVWWQEFD--VTPRQLEEVS 260
+ L T+ LQ+ P +A I LA K+ ++P S K WW+ D VT L+E++
Sbjct: 179 TNSLHLTTFSLQYTPPVVACVCIHLACKWSNWEIPVSTDGKHWWEYVDATVTLELLDELT 238
Query: 261 NQMLELYEQ--NRMAQSNDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPSSTLETS 318
++ L++ E+ NR+ + + + T DE S L++ SS+ T
Sbjct: 239 HEFLQILEKTPNRLKRIRNWRACQAA--KKTKADDRGTDENTSEQTILNMISQSSSDTTI 296
Query: 319 KPATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPVEDDVKGNLHP-KQEPSAYEENMQEA 377
S + +++ P + S + V+ + + P K EP+ +
Sbjct: 297 AGLMSMSTSSTTSTVPSLPTTEESSSNLSGVEMLQGERWLSSQPPFKLEPAQGHRTSENL 356
Query: 378 QDMVRSRSGYAKG-NQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQESNVGRSNIKEEDV 436
+ S G N Q+ ++ + + R+ + +E + + E +V
Sbjct: 357 ALIGVDHSLQQDGSNAFISQKQNSSKSVPSAKVSLKEYRAKHAEELAAQKRQLENMEANV 416
Query: 437 ELNDKHSSKNLNHRDATFASPNLEGIKKIDRDKVKAALEKRKKATGNISKKTELVDDEDL 496
+ ++++NL + +S L+ + + + LEK K T + D+
Sbjct: 417 KSQYAYAAQNLLSHHDSHSSVILKMPIEGSENPERPFLEKPDK-TALKMRIPVASGDKAA 475
Query: 497 IERVRELEDGIELAAQSEKNKQDGRKSLSKSLDRSAFENMQH-GKHQDHSEEHLH 550
+ E++ I++ A +K+ S+D S ++ +H KH+ H H H
Sbjct: 476 SSKPEEIKMRIKVHAAPDKH---------NSIDDSVTKSREHKEKHKTHPSNHHH 521
>CCT_DROME (O96433) Cyclin T
Length = 1097
Score = 125 bits (314), Expect = 3e-28
Identities = 81/270 (30%), Positives = 144/270 (53%), Gaps = 15/270 (5%)
Query: 33 WYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFF 92
WYFS ++ NSPS++ G+ E R+ +Q++G RL+V Q+ I TAI++ HRF+
Sbjct: 45 WYFSNDQLA-NSPSRRCGIKGDDELQYRQMTAYLIQEMGQRLQVSQLCINTAIVYMHRFY 103
Query: 93 LRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYE 152
S R +A+ +FLA KVEE PR L+ VI + + + P Q Y
Sbjct: 104 AFHSFTHFHRNSMASASLFLAAKVEEQPRKLEHVIRAANKCL---PPTTEQN------YA 154
Query: 153 QQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLR-TS 211
+ + ++ E V+L TLGFD+ + HP+ +V + K ++ LAQ ++ ++ L TS
Sbjct: 155 ELAQELVFNENVLLQTLGFDVAIDHPHTHVVRTCQLVKACKD-LAQTSYFLASNSLHLTS 213
Query: 212 LCLQFKPHHIAAGAIFLAAKFLKVKLPSDGE-KVWWQEFD--VTPRQLEEVSNQMLELYE 268
+CLQ++P +A I+LA K+ + ++P E K W+ D V+ L++++++ + +YE
Sbjct: 214 MCLQYRPTVVACFCIYLACKWSRWEIPQSTEGKHWFYYVDKTVSLDLLKQLTDEFIAIYE 273
Query: 269 QNRMAQSNDVEGTTGGGNRATPKAPTSNDE 298
++ + + A+ + S D+
Sbjct: 274 KSPARLKSKLNSIKAIAQGASNRTANSKDK 303
>CG1C_DROME (P25008) G1/S-specific cyclin C
Length = 267
Score = 91.3 bits (225), Expect = 7e-18
Identities = 65/214 (30%), Positives = 104/214 (48%), Gaps = 25/214 (11%)
Query: 63 YCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRP 122
+ +Q LG +LK+ Q IATA ++ RF+ R S D ++A C+ LA KVEE
Sbjct: 44 FANVIQVLGEQLKLRQQVIATATVYFKRFYARNSLKNIDPLLLAPTCILLASKVEE---- 99
Query: 123 LKDVILVSYEIIHKKDPAAV--QRIKQKDVYEQQKEL------ILLGERVVLATLGFDLN 174
+ +I ++ IK K Y +E IL E +L L L
Sbjct: 100 --------FGVISNSRLISICQSAIKTKFSYAYAQEFPYRTNHILECEFYLLENLDCCLI 151
Query: 175 VHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLK 234
V+ PY+PL++ ++ ++ L ++W VND LRT +CL + P+ IA + +A
Sbjct: 152 VYQPYRPLLQLVQDMG-QEDQLLTLSWRIVNDSLRTDVCLLYPPYQIAIACLQIAC---- 206
Query: 235 VKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYE 268
V L D K W+ E +V +++E+ ++ LYE
Sbjct: 207 VILQKDATKQWFAELNVDLDKVQEIVRAIVNLYE 240
>CG1C_CHICK (P55168) Cyclin C
Length = 283
Score = 90.9 bits (224), Expect = 9e-18
Identities = 73/246 (29%), Positives = 119/246 (47%), Gaps = 26/246 (10%)
Query: 32 RWYFSRKEIEENSPSKQDGVDLKKEAY--LRKSYCTFLQDLGMRLKVPQVTIATAIIFCH 89
+W ++++ + ++D L +E Y L+ + +Q LG LK+ Q IATA ++
Sbjct: 13 QWILDKQDLLKER--QKDLKFLSEEEYWKLQIFFTNVIQALGEHLKLRQQVIATATVYFK 70
Query: 90 RFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKD 149
RF+ R S D ++A C+FLA KVEE + + L+S AA +K +
Sbjct: 71 RFYARYSLKSIDPVLMAPTCVFLASKVEEF-GVVSNTRLIS---------AATSVLKTRF 120
Query: 150 VYEQQKEL------ILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNF 203
Y KE IL E +L + L V+HPY+PL++ ++ ++ L +AW
Sbjct: 121 SYAFPKEFPYRMNHILECEFYLLELMDCCLIVYHPYRPLLQYVQDMG-QEDMLLPLAWRI 179
Query: 204 VNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQM 263
VND RT LCL + P IA + +A + + W+ E V ++ E+ +
Sbjct: 180 VNDTYRTDLCLLYPPFMIALACLHVAC-----VVQQKDARQWFAELSVDMEKILEIIRVI 234
Query: 264 LELYEQ 269
L+LYEQ
Sbjct: 235 LKLYEQ 240
>CG1C_HUMAN (P24863) Cyclin C
Length = 303
Score = 90.5 bits (223), Expect = 1e-17
Identities = 74/259 (28%), Positives = 123/259 (46%), Gaps = 26/259 (10%)
Query: 19 SQGGSQDNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAY--LRKSYCTFLQDLGMRLKV 76
S G+ + +W ++++ + ++D L +E Y L+ + +Q LG LK+
Sbjct: 20 SMAGNFWQSSHYLQWILDKQDLLKER--QKDLKFLSEEEYWKLQIFFTNVIQALGEHLKL 77
Query: 77 PQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHK 136
Q IATA ++ RF+ R S D ++A C+FLA KVEE ++ + +I
Sbjct: 78 RQQVIATATVYFKRFYARYSLKSIDPVLMAPTCVFLASKVEEFG------VVSNTRLI-- 129
Query: 137 KDPAAVQRIKQKDVYEQQKEL------ILLGERVVLATLGFDLNVHHPYKPLVEAIKKFK 190
AA +K + Y KE IL E +L + L V+HPY+PL++ ++
Sbjct: 130 --AAATSVLKTRFSYAFPKEFPYRMNHILECEFYLLELMDCCLIVYHPYRPLLQYVQDMG 187
Query: 191 VAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFD 250
++ L +AW VND RT LCL + P IA + +A + + W+ E
Sbjct: 188 -QEDMLLPLAWRIVNDTYRTDLCLLYPPFMIALACLHVAC-----VVQQKDARQWFAELS 241
Query: 251 VTPRQLEEVSNQMLELYEQ 269
V ++ E+ +L+LYEQ
Sbjct: 242 VDMEKILEIIRVILKLYEQ 260
>YL33_CAEEL (P34424) Hypothetical protein F44B9.3 in chromosome III
Length = 555
Score = 90.1 bits (222), Expect = 2e-17
Identities = 94/406 (23%), Positives = 177/406 (43%), Gaps = 40/406 (9%)
Query: 33 WYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDLGMRL---KVPQVTIATAIIFCH 89
W ++++ ++PS+++G+ ++E R+ F+ D+ M+L K A +
Sbjct: 23 WLRKKQDMLADTPSRREGMTYEEELSKRQQGGVFIFDIAMQLTHGKGEHGLSGVAATLFN 82
Query: 90 RFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVI--LVSYEIIHKKDPAAVQRIKQ 147
RFF S + D R +A C+FLAGK E+ P+ LK V+ L ++ H K + +
Sbjct: 83 RFFNVHSLKRCDFRDVAAACVFLAGKNEDAPKKLKYVVTQLWQFKYPHNK------QFQS 136
Query: 148 KDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNA---LAQVAWNFV 204
+ + Q ++ L E V+L T+ FD+NV P++ +++ ++ + +N + + A+
Sbjct: 137 EQHFLDQCNVVTLIEDVLLKTISFDINVDLPHQYVLKLMRDVEKGRNVYKDMVKTAYYMA 196
Query: 205 NDGL-RTSLCLQFKPHHIAAGAIFLAAKFLKVKL----PSDGEKVWW--QEFDVTPRQLE 257
D L T +++ IA + +AA F + + P + W+ ++ +T ++E
Sbjct: 197 TDVLIITDWSVRYSCASIATACVNIAAFFHNINMDDIVPFELSDRWYRLEDQSMTREEVE 256
Query: 258 EVSNQMLELYEQNRMAQSNDVEGTTGGGNRATPKAPTSNDEAASTNRNLHIGGPSSTLET 317
++ + L+++ +N ++ G P + + G SS L
Sbjct: 257 AMTKEFLDIFSRNPQFHIGSLKKIDPLGKVKIVGMPPQVSSTVTPS------GSSSNL-- 308
Query: 318 SKPATSKNVFVSSANHVGRPVSNHGVSGSTEVKHPVEDDVKGNLHPKQEPSAYEENMQEA 377
K + + S +P+SN S ST + DVK +QE E+ M+EA
Sbjct: 309 ------KKIDLESYKGRPKPLSNSSDSPST--RPSFLPDVKNQKVVEQE--LMEQRMKEA 358
Query: 378 QDMVRSRSGY-AKGNQHPKQEPSTYEEKMQEAQDMVRSRSGYGKEQ 422
R+G+ + HP ST + RS SG Q
Sbjct: 359 AQQKIHRNGHRPSSSSHPLHHHSTSSSASNNSNHQNRSSSGLSAHQ 404
>CG1C_RAT (P39947) Cyclin C
Length = 298
Score = 89.7 bits (221), Expect = 2e-17
Identities = 69/225 (30%), Positives = 108/225 (47%), Gaps = 24/225 (10%)
Query: 53 LKKEAY--LRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCM 110
L +E Y L+ + +Q LG LK+ Q IATA ++ RF+ R S D ++A C+
Sbjct: 47 LSEEEYWKLQIFFTNVIQALGEHLKLRQQVIATATVYFKRFYARYSLKSIDPVLMAPTCV 106
Query: 111 FLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKEL------ILLGERV 164
FLA KVEE ++ + +I A +K + Y KE IL E
Sbjct: 107 FLASKVEEFG------VVSNTSLI----AATTSVLKTRFSYASPKEFPYRMNHILECEFY 156
Query: 165 VLATLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAG 224
+L + L V+HPY+PL++ ++ ++ L +AW VND RT LCL + P IA
Sbjct: 157 LLELMDCCLIVYHPYRPLLQYVQDMG-QEDVLLPLAWRIVNDTYRTDLCLLYPPFMIALA 215
Query: 225 AIFLAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYEQ 269
+ +A + + W+ E V ++ E+ +L+LYEQ
Sbjct: 216 CLHVAC-----VVQQKDARQWFAELSVDMEKILEIIRVILKLYEQ 255
>YL34_CAEEL (P34425) Hypothetical protein F44B9.4 in chromosome III
Length = 468
Score = 89.4 bits (220), Expect = 3e-17
Identities = 68/259 (26%), Positives = 127/259 (48%), Gaps = 26/259 (10%)
Query: 31 SRWYFSRKEIEENSPSKQDGVDLKKEAYLRKSYCTFLQDL--------GMRLKVPQVTIA 82
++W F+++E+++ + S Q+G+ ++E R+ F+Q++ ++K+ +
Sbjct: 18 TKWLFTKEEMKKTA-SIQEGMSREEELASRQMAAAFIQEMIDGLNNVKDPKMKIGHTGLC 76
Query: 83 TAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAV 142
A HRF+ S K D R + C+FLAGK +E PR L VI V E +KD
Sbjct: 77 VAHTHMHRFYYLHSFKKYDYRDVGAACVFLAGKSQECPRKLSHVISVWRE---RKDR--- 130
Query: 143 QRIKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEAIKKF--KVAQNALAQVA 200
+++ + + ++I+L E ++L T+ FDLNVH P+ +++ +KK K L A
Sbjct: 131 KQLTTETARNEAAQIIVLLESMILQTIAFDLNVHLPHIYVLDIMKKVDKKEHYRPLTSCA 190
Query: 201 WNFVNDGLR-TSLCLQFKPHHIAAGAIFLAAKFLKVKLPS------DGEKVWWQEFD--V 251
+ F D + T L++ ++ I L A + V++ + + W+ +FD +
Sbjct: 191 YYFATDVIAVTDWSLRYSAASMSIVIIHLMAAYANVRIERLFADFINEDSPWYAKFDETM 250
Query: 252 TPRQLEEVSNQMLELYEQN 270
T +L E+ L Y +
Sbjct: 251 TNEKLREMEVDFLVTYRNS 269
>CG1C_ORYSA (P93411) G1/S-specific cyclin C-type
Length = 257
Score = 88.2 bits (217), Expect = 6e-17
Identities = 64/221 (28%), Positives = 114/221 (50%), Gaps = 22/221 (9%)
Query: 50 GVDLKKEAYLRKSYCTFLQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKNDRRIIATVC 109
G+ L++ ++ + L ++KV Q IATA+ + R + R+S + D R++A C
Sbjct: 31 GITLEEFRLVKIHMSFHIWRLAQQVKVRQRVIATAVTYFRRVYTRKSMTEYDPRLVAPTC 90
Query: 110 MFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQ--RIKQKDVYEQQKELILLGERVVLA 167
++LA KVEE+ ++ +LV Y KK A+ + R + KD+ E + +L L
Sbjct: 91 LYLASKVEES--TVQARLLVFY---IKKMCASDEKYRFEIKDILEMEMKL--------LE 137
Query: 168 TLGFDLNVHHPYKPLVEAIKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIF 227
L + L V+HPY+PL++ ++ + L Q AW VND + L L P+ IA I+
Sbjct: 138 ALDYYLVVYHPYRPLLQLLQDAGITD--LTQFAWGIVNDTYKMDLILIHPPYMIALACIY 195
Query: 228 LAAKFLKVKLPSDGEKVWWQEFDVTPRQLEEVSNQMLELYE 268
+A+ L +W++E V ++ +S ++L+ Y+
Sbjct: 196 IAS-----VLKDKDITLWFEELRVDMNIVKNISMEILDFYD 231
>CG1C_MOUSE (Q62447) Cyclin C
Length = 304
Score = 87.0 bits (214), Expect = 1e-16
Identities = 73/259 (28%), Positives = 121/259 (46%), Gaps = 26/259 (10%)
Query: 19 SQGGSQDNQEYGSRWYFSRKEIEENSPSKQDGVDLKKEAY--LRKSYCTFLQDLGMRLKV 76
S G+ + +W ++++ + ++D L +E Y L+ + +Q LG LK+
Sbjct: 21 SMAGNFWQSSHYLQWILDKQDLLKER--QKDLKFLSEEEYWKLQIFFTNVIQALGEHLKL 78
Query: 77 PQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHK 136
Q IATA ++ RF R S D ++A C+FLA KVEE ++ + +I
Sbjct: 79 RQQVIATATVYFKRFDARYSLKSIDPVLMAPTCVFLASKVEEFG------VVSNTRLI-- 130
Query: 137 KDPAAVQRIKQKDVYEQQKEL------ILLGERVVLATLGFDLNVHHPYKPLVEAIKKFK 190
A +K + Y KE IL E +L + L V+HPY+PL++ ++
Sbjct: 131 --AATTSVLKTRFSYAFPKEFPYRMNHILECEFYLLELMDCCLIVYHPYRPLLQYVQDMG 188
Query: 191 VAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSDGEKVWWQEFD 250
++ L +AW VND RT LCL + P IA + +A + + W+ E
Sbjct: 189 -QEDVLLPLAWRIVNDTYRTDLCLLYPPFMIALACLHVAC-----VVQQKDARQWFAELS 242
Query: 251 VTPRQLEEVSNQMLELYEQ 269
V ++ E+ +L+LYEQ
Sbjct: 243 VDMEKILEIIRVILKLYEQ 261
>UME3_YEAST (P47821) RNA polymerase II holoenzyme cyclin-like
subunit
Length = 323
Score = 72.0 bits (175), Expect = 4e-12
Identities = 53/198 (26%), Positives = 92/198 (45%), Gaps = 21/198 (10%)
Query: 38 KEIEENSPSKQDGVDLKKEAYLRKSYCTFL-QDLGMRLKVPQVTIATAIIFCHRFFLRQS 96
+ I +N P + K+ LR YC FL LG RL + Q +ATA I+ RF ++ S
Sbjct: 53 QSITKNIPITHRDLHYDKDYNLR-IYCYFLIMKLGRRLNIRQYALATAHIYLSRFLIKAS 111
Query: 97 HAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIHKKDPAAVQRIKQKDVYEQQKE 156
+ + ++ T C++LA KVEE P+ ++ ++ + + + P ++ + + Y
Sbjct: 112 VREINLYMLVTTCVYLACKVEECPQYIRTLVSEARTLWPEFIPPDPTKVTEFEFY----- 166
Query: 157 LILLGERVVLATLGFDLNVHHPYKPLVEAIKKFKVAQNALA------QVAWNFVNDGLRT 210
+L L L VHHPY+ L + ++ K + Q W+ +ND
Sbjct: 167 --------LLEELESYLIVHHPYQSLKQIVQVLKQPPFQITLSSDDLQNCWSLINDSYIN 218
Query: 211 SLCLQFKPHHIAAGAIFL 228
+ L + PH IA +F+
Sbjct: 219 DVHLLYPPHIIAVACLFI 236
>SRBB_SCHPO (O94503) RNA polymerase II holoenzyme cyclin-like
subunit (Suppressor of RNA polymerase B srb11)
Length = 228
Score = 71.6 bits (174), Expect = 6e-12
Identities = 47/164 (28%), Positives = 83/164 (49%), Gaps = 15/164 (9%)
Query: 67 LQDLGMRLKVPQVTIATAIIFCHRFFLRQSHAKN-DRRIIATVCMFLAGKVEETPRPLKD 125
+Q G RL++ Q +ATAI+ R+ L+++ K + C++L+ KVEE P ++
Sbjct: 39 VQTFGDRLRLRQRVLATAIVLLRRYMLKKNEEKGFSLEALVATCIYLSCKVEECPVHIRT 98
Query: 126 VILVSYEIIHKKDPAAVQRIKQKDVYEQQKELILLGERVVLATLGFDLNVHHPYKPLVEA 185
+ + ++ K ++ + ++ E + E+I + L L VHHPY L +A
Sbjct: 99 ICNEANDLWSLK-----VKLSRSNISEIEFEII--------SVLDAFLIVHHPYTSLEQA 145
Query: 186 IKKFKVAQNALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLA 229
+ Q L + AW+ VND +SLCL PH +A A+ ++
Sbjct: 146 FHDGIINQKQL-EFAWSIVNDSYASSLCLMAHPHQLAYAALLIS 188
>CYCH_HUMAN (P51946) Cyclin H (MO15-associated protein) (p37) (p34)
Length = 323
Score = 60.5 bits (145), Expect = 1e-08
Identities = 55/211 (26%), Positives = 95/211 (44%), Gaps = 23/211 (10%)
Query: 76 VPQVTIATAIIFCHRFFLRQSHAKNDRRIIATVCMFLAGKVEETPRPLKDVILVSYEIIH 135
+P+ + TA ++ RF+L S + RII C FLA KV+E +
Sbjct: 75 MPRSVVGTACMYFKRFYLNNSVMEYHPRIIMLTCAFLACKVDE---------------FN 119
Query: 136 KKDPAAVQRIKQKDV-YEQQKELILLGERVVLATLGFDLNVHHPYKP----LVEAIKKFK 190
P V +++ + E+ E IL E +++ L F L VH+PY+P L++ ++
Sbjct: 120 VSSPQFVGNLRESPLGQEKALEQILEYELLLIQQLNFHLIVHNPYRPFEGFLIDLKTRYP 179
Query: 191 VAQN--ALAQVAWNFVNDGLRTSLCLQFKPHHIAAGAIFLAAKFLKVKLPSD-GEKVWWQ 247
+ +N L + A +F+N T L + P IA AI +A + + S E + +
Sbjct: 180 ILENPEILRKTADDFLNRIALTDAYLLYTPSQIALTAILSSASRAGITMESYLSESLMLK 239
Query: 248 EFDVTPRQLEEVSNQMLELYEQNRMAQSNDV 278
E QL ++ M L ++ +S +V
Sbjct: 240 ENRTCLSQLLDIMKSMRNLVKKYEPPRSEEV 270
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 77,277,927
Number of Sequences: 164201
Number of extensions: 3427353
Number of successful extensions: 10300
Number of sequences better than 10.0: 374
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 340
Number of HSP's that attempted gapping in prelim test: 9853
Number of HSP's gapped (non-prelim): 653
length of query: 641
length of database: 59,974,054
effective HSP length: 117
effective length of query: 524
effective length of database: 40,762,537
effective search space: 21359569388
effective search space used: 21359569388
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0176a.14


