

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
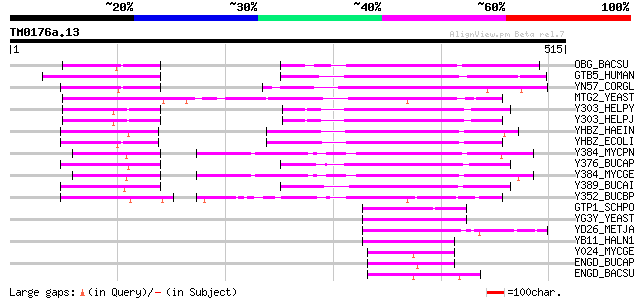
Query= TM0176a.13
(515 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
OBG_BACSU (P20964) Spo0B-associated GTP-binding protein 176 1e-43
GTB5_HUMAN (Q9H4K7) Putative GTP-binding protein 5 173 1e-42
YN57_CORGL (P46584) Probable GTP-binding protein Cgl2357/cg2589 161 3e-39
MTG2_YEAST (P38860) Mitochondrial GTPase 2 150 1e-35
Y303_HELPY (O25074) Probable GTP-binding protein HP0303 140 6e-33
Y303_HELPJ (Q9ZMD3) Probable GTP-binding protein JHP0288 140 8e-33
YHBZ_HAEIN (P44915) Hypothetical GTP-binding protein HI0877 137 5e-32
YHBZ_ECOLI (P42641) Hypothetical GTP-binding protein yhbZ 137 9e-32
Y384_MYCPN (P75215) Probable GTP-binding protein MG384 homolog 128 4e-29
Y376_BUCAP (Q8K9G1) Hypothetical GTP-binding protein BUsg376 125 3e-28
Y384_MYCGE (P47624) Probable GTP-binding protein MG384 119 1e-26
Y389_BUCAI (P57469) Hypothetical GTP-binding protein BU389 118 4e-26
Y352_BUCBP (Q89AE7) Hypothetical GTP-binding protein bbp352 108 3e-23
GTP1_SCHPO (P32235) GTP-binding protein 1 69 3e-11
YG3Y_YEAST (P53295) Hypothetical 41.0 kDa protein in YIP1-CBP4 i... 69 4e-11
YD26_METJA (Q58722) Hypothetical GTP-binding protein MJ1326 66 2e-10
YB11_HALN1 (P17103) Hypothetical protein Vng1111g 65 3e-10
Y024_MYCGE (P47270) Probable GTP-binding protein MG024 65 3e-10
ENGD_BUCAP (Q8K9V2) GTP-dependent nucleic acid-binding protein engD 65 4e-10
ENGD_BACSU (P37518) GTP-dependent nucleic acid-binding protein engD 63 2e-09
>OBG_BACSU (P20964) Spo0B-associated GTP-binding protein
Length = 428
Score = 176 bits (446), Expect = 1e-43
Identities = 103/242 (42%), Positives = 139/242 (56%), Gaps = 27/242 (11%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNA 311
+A+LTE GQ+ ++ARGG GG GN T A Q N
Sbjct: 104 IADLTEHGQRAVIARGGRGGRGN--------SRFATPANPAPQLSEN------------- 142
Query: 312 GMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNF 371
G PG E ++LELK +ADV VG P+ GKSTLL +S AKP + Y FTTL PNLG +
Sbjct: 143 GEPGKERYIVLELKVLADVGLVGFPSVGKSTLLSVVSSAKPKIADYHFTTLVPNLGMVET 202
Query: 372 DD-FSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
DD S +AD+PGLI+GAHQ GLGH FLRHIERT+V+ +V+D++ G +G P++
Sbjct: 203 DDGRSFVMADLPGLIEGAHQGVGLGHQFLRHIERTRVIVHVIDMS----GLEGRDPYDDY 258
Query: 431 KDLILELEYHQDGLSNRPSLIVANKID-EEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
+ EL + L+ RP +IVANK+D E A+ + ++ P+F + AV EG+
Sbjct: 259 LTINQELSEYNLRLTERPQIIVANKMDMPEAAENLEAFKEKLTDDYPVFPISAVTREGLR 318
Query: 490 EL 491
EL
Sbjct: 319 EL 320
Score = 69.3 bits (168), Expect = 2e-11
Identities = 37/94 (39%), Positives = 54/94 (57%), Gaps = 4/94 (4%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSP---RVWDFSGLQ 106
+D+ K+ K GDGG+G +F R ++ +G GG+GG+GGDV+ E + DF +
Sbjct: 3 VDQVKVYVKGGDGGNGMVAFRREKYVPKGGPAGGDGGKGGDVVFEVDEGLRTLMDFR-YK 61
Query: 107 HHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
H A +G G SKN+ G D V+ VP GTV+
Sbjct: 62 KHFKAIRGEHGMSKNQHGRNADDMVIKVPPGTVV 95
>GTB5_HUMAN (Q9H4K7) Putative GTP-binding protein 5
Length = 406
Score = 173 bits (438), Expect = 1e-42
Identities = 107/249 (42%), Positives = 144/249 (56%), Gaps = 32/249 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGN-LSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLN 310
VA+L+ G + I A GG GG GN + ++R P+T
Sbjct: 170 VADLSCVGDEYIAALGGAGGKGNRFFLANNNRAPVTC----------------------T 207
Query: 311 AGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLN 370
G PG + VL LELK++A VG PNAGKS+LL AIS A+PAV Y FTTL+P++G ++
Sbjct: 208 PGQPGQQRVLHLELKTVAHAGMVGFPNAGKSSLLRAISNARPAVASYPFTTLKPHVGIVH 267
Query: 371 FD-DFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQ 429
++ I VADIPG+I+GAHQNRGLG AFLRHIER + L +VVDL+ PW Q
Sbjct: 268 YEGHLQIAVADIPGIIRGAHQNRGLGSAFLRHIERCRFLLFVVDLSQP-------EPWTQ 320
Query: 430 LKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
+ DL ELE ++ GLS RP IVANKID A +L+ + G + + A+ E +
Sbjct: 321 VDDLKYELEMYEKGLSARPHAIVANKIDLPEAQANLSQLRDHL-GQEVIVLSALTGENLE 379
Query: 490 ELKAGLRML 498
+L L++L
Sbjct: 380 QLLLHLKVL 388
Score = 69.3 bits (168), Expect = 2e-11
Identities = 38/110 (34%), Positives = 53/110 (47%)
Query: 31 YSDAPHKKSKLAPLQERRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGD 90
+ + P KK +R +D +++ G+GG+G S FH G DGG+GG GG
Sbjct: 54 HQELPGKKLLSEKKLKRYFVDYRRVLVCGGNGGAGASCFHSEPRKEFGGPDGGDGGNGGH 113
Query: 91 VILECSPRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
VIL +V S + G G SKN G GA + VPVGT++
Sbjct: 114 VILRVDQQVKSLSSVLSRYQGFSGEDGGSKNCFGRSGAVLYIRVPVGTLV 163
>YN57_CORGL (P46584) Probable GTP-binding protein Cgl2357/cg2589
Length = 501
Score = 161 bits (408), Expect = 3e-39
Identities = 108/274 (39%), Positives = 144/274 (52%), Gaps = 36/274 (13%)
Query: 235 GTEYIGEEQEEQEILYNVAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQ 294
GT + E+ E +A+LT G + I A GG GG GN + + +RK
Sbjct: 94 GTVVLNEKGE------TLADLTSVGMKFIAAAGGNGGLGNAALASKARK----------- 136
Query: 295 HISNLEDSDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAV 354
G PG + LILELKS+ADV VG P+AGKS+L+ +S AKP +
Sbjct: 137 ----------APGFALIGEPGEAHDLILELKSMADVGLVGFPSAGKSSLISVMSAAKPKI 186
Query: 355 GHYAFTTLRPNLGNLNFDDFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDL 414
G Y FTTL+PNLG +N + T+AD+PGLI GA + +GLG FLRHIERT VL +VVD
Sbjct: 187 GDYPFTTLQPNLGVVNVGHETFTMADVPGLIPGASEGKGLGLDFLRHIERTSVLVHVVDT 246
Query: 415 AAALHGRKGIPPWEQLKDLILELEYHQD------GLSNRPSLIVANKIDEEGADEVYLEL 468
A GR I E L+ + + D LS RP L+V NK D A+E+ L
Sbjct: 247 ATMDPGRDPISDIEALEAELAAYQSALDEDTGLGDLSQRPRLVVLNKADVPEAEELAEFL 306
Query: 469 QRRVH---GVPIFRVCAVLEEGIPELKAGLRMLV 499
+ + G P+F + AV +G+ LK L +V
Sbjct: 307 KEDIEKQFGWPVFIISAVARKGLDPLKYKLLEIV 340
Score = 79.7 bits (195), Expect = 2e-14
Identities = 43/96 (44%), Positives = 57/96 (58%), Gaps = 4/96 (4%)
Query: 48 RMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRV---WDFSG 104
R ID+ + AGDGG+GC S HR + G DGGNGG GGD+ILE + +V DF
Sbjct: 3 RFIDRVVLHLAAGDGGNGCVSVHREKFKPLGGPDGGNGGHGGDIILEVTAQVHTLLDFH- 61
Query: 105 LQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
H+ A +G G+ ++ G RG D V+ VP GTV+
Sbjct: 62 FHPHVKAERGANGAGDHRNGARGKDLVLEVPPGTVV 97
>MTG2_YEAST (P38860) Mitochondrial GTPase 2
Length = 499
Score = 150 bits (378), Expect = 1e-35
Identities = 125/415 (30%), Positives = 194/415 (46%), Gaps = 50/415 (12%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQHHL 109
+D + K+G GGSG SF R G DGG+GG GG V ++ + + ++
Sbjct: 92 VDVRIVKCKSGAGGSGAVSFFRDAGRSIGPPDGGDGGAGGSVYIQAVAGLGSLAKMKTTY 151
Query: 110 IAGKGGPGSSKNKIGTRGADKVVHVPVGTVIH--LVNGDIPSIVKTESSADTDPW--EIS 165
A G G+++ G RG D ++ VPVGTV+ L + +V+ E D + I
Sbjct: 152 TAEDGEAGAARQLDGMRGRDVLIQVPVGTVVKWCLPPQKVRELVEREMRKDNNATLRSIL 211
Query: 166 GALVDDLPDHGNGSISNATNGEEVKTIHPTGCSSSQAAEKNVEKSVKSGHVASTDVLSQL 225
G+ +L S+S+ ++ ++++ + AE + K + + D L
Sbjct: 212 GSTAVNL------SVSSGSHRKKIQLYR------HEMAESWLFKDKAKEYHENKDWFKDL 259
Query: 226 SISNGTPKFGTEYIGEEQEEQEILYNVAELTEEGQQMIVARGGEGGPGNLSTSKDS-RKP 284
E E +Q L + + + + +GG+GG GN+ + R P
Sbjct: 260 HKKMEAYDHSLEQ-SELFNDQFPLAGLDLNQPMTKPVCLLKGGQGGLGNMHFLTNLIRNP 318
Query: 285 ITTKAGACRQHISNLEDSDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLL 344
+K G G E + ELKSIAD+ +G+PNAGKST+L
Sbjct: 319 RFSKPGR----------------------NGLEQHFLFELKSIADLGLIGLPNAGKSTIL 356
Query: 345 GAISRAKPAVGHYAFTTLRPNLG--NLNFDDFSITVADIPGLIKGAHQNRGLGHAFLRHI 402
IS AKP +GH+ FTTL P +G +L F TVADIPG+I+GA ++G+G FLRHI
Sbjct: 357 NKISNAKPKIGHWQFTTLSPTIGTVSLGFGQDVFTVADIPGIIQGASLDKGMGLEFLRHI 416
Query: 403 ERTKVLAYVVDLAAALHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANKID 457
ER+ +V+DL+ P L+ L++E + + + LIV NK+D
Sbjct: 417 ERSNGWVFVLDLS-------NKNPLNDLQ-LLIEEVGTLEKVKTKNILIVCNKVD 463
>Y303_HELPY (O25074) Probable GTP-binding protein HP0303
Length = 360
Score = 140 bits (354), Expect = 6e-33
Identities = 90/213 (42%), Positives = 124/213 (57%), Gaps = 28/213 (13%)
Query: 254 ELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNAGM 313
+L E ++++ +GG+GG GN + K + K T A G+
Sbjct: 104 DLVEPKERVLALKGGKGGLGN-AHFKSATKQQPTYA--------------------QKGL 142
Query: 314 PGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDD 373
G E + LELK IAD+ VG PNAGKSTL+ IS AKP + +Y FTTL PNLG ++ D+
Sbjct: 143 EGVEKCVRLELKLIADIGLVGFPNAGKSTLISTISNAKPKIANYEFTTLVPNLGVVSVDE 202
Query: 374 FS-ITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQLKD 432
S +ADIPG+I+GA + +GLG +FL+HIERTKVLA+V+D + G K EQ +
Sbjct: 203 KSGFLMADIPGIIEGASEGKGLGISFLKHIERTKVLAFVLDASRLDLGIK-----EQYQR 257
Query: 433 LILELEYHQDGLSNRPSLIVANKID-EEGADEV 464
L LELE L+N+P ++ NK D E DE+
Sbjct: 258 LRLELEKFSSALANKPFGVLLNKCDVVENIDEM 290
Score = 70.9 bits (172), Expect = 8e-12
Identities = 38/94 (40%), Positives = 53/94 (55%), Gaps = 4/94 (4%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILEC---SPRVWDFSGLQ 106
+D +II +G GG G SF R + +G DGG+GG GGDV E + + F G +
Sbjct: 3 VDSVEIIIASGKGGPGMVSFRREKFVIKGGPDGGDGGDGGDVYFEVDNNTDTLASFRGTK 62
Query: 107 HHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
HH A G PG ++N G +G DK++ VP GT +
Sbjct: 63 HHK-AKNGAPGGTRNCAGKKGEDKIIVVPPGTQV 95
>Y303_HELPJ (Q9ZMD3) Probable GTP-binding protein JHP0288
Length = 360
Score = 140 bits (353), Expect = 8e-33
Identities = 88/205 (42%), Positives = 120/205 (57%), Gaps = 27/205 (13%)
Query: 254 ELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNAGM 313
+L E ++++ +GG+GG GN + K + K T A G+
Sbjct: 104 DLVEPKERVLALKGGKGGLGN-AHFKSATKQQPTYA--------------------QKGL 142
Query: 314 PGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDD 373
G E + LELK IAD+ VG PNAGKSTL+ IS AKP + +Y FTTL PNLG ++ D+
Sbjct: 143 EGVEKCVRLELKLIADIGLVGFPNAGKSTLISTISNAKPKIANYEFTTLVPNLGVVSVDE 202
Query: 374 FS-ITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQLKD 432
S +ADIPG+I+GA + +GLG +FL+HIERTKVLA+V+D + G K EQ K
Sbjct: 203 KSEFLMADIPGIIEGASEGKGLGISFLKHIERTKVLAFVLDASRLDLGIK-----EQYKR 257
Query: 433 LILELEYHQDGLSNRPSLIVANKID 457
L LELE L+N+P ++ NK D
Sbjct: 258 LRLELEKFSPTLANKPFGVLLNKCD 282
Score = 70.9 bits (172), Expect = 8e-12
Identities = 38/94 (40%), Positives = 53/94 (55%), Gaps = 4/94 (4%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILEC---SPRVWDFSGLQ 106
+D +II +G GG G SF R + +G DGG+GG GGDV E + + F G +
Sbjct: 3 VDSVEIIIASGKGGPGMVSFRREKFVIKGGPDGGDGGDGGDVYFEVDNNTDTLASFRGTK 62
Query: 107 HHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
HH A G PG ++N G +G DK++ VP GT +
Sbjct: 63 HHK-AKNGAPGGTRNCAGKKGEDKIIVVPPGTQV 95
>YHBZ_HAEIN (P44915) Hypothetical GTP-binding protein HI0877
Length = 390
Score = 137 bits (346), Expect = 5e-32
Identities = 88/238 (36%), Positives = 126/238 (51%), Gaps = 29/238 (12%)
Query: 239 IGEEQEEQEILYNVAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISN 298
+G + + + +LT+ GQ+M+VA+GG G GN + K
Sbjct: 92 VGTRAIDNDTKETLGDLTQHGQKMLVAKGGYHGLGNTRFKSSVNRAPRQKT--------- 142
Query: 299 LEDSDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYA 358
G PG + L+LEL +ADV +G+PNAGKST + A+S AKP V Y
Sbjct: 143 ------------MGTPGEKRDLLLELMLLADVGMLGLPNAGKSTFIRAVSAAKPKVADYP 190
Query: 359 FTTLRPNLGNLNFDD-FSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAA 417
FTTL P+LG + DD S VADIPGLI+GA GLG FL+H+ER +VL ++VD+A
Sbjct: 191 FTTLVPSLGVVKVDDSHSFVVADIPGLIEGAADGAGLGIRFLKHLERCRVLIHLVDIAPI 250
Query: 418 LHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANKID---EEGADEVYLELQRRV 472
G P + + + EL + + LS +P +V NKID +E A+E E+ ++
Sbjct: 251 ----DGSNPADNMAIIESELFQYSEKLSEKPRWLVFNKIDTMSDEEAEERAREIAEQL 304
Score = 65.9 bits (159), Expect = 2e-10
Identities = 37/93 (39%), Positives = 52/93 (55%), Gaps = 2/93 (2%)
Query: 48 RMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH 107
+ ID+ I +AGDGG+GC SF R + +G DGG+GG GGDV L+ + +
Sbjct: 2 KFIDESLIRIEAGDGGNGCVSFRREKFIPKGGPDGGDGGDGGDVYLQADENLNTLIDYRF 61
Query: 108 H--LIAGKGGPGSSKNKIGTRGADKVVHVPVGT 138
+ A +G G S + G RG D ++ VPVGT
Sbjct: 62 NKRFAAERGENGRSSDCTGRRGKDIILPVPVGT 94
>YHBZ_ECOLI (P42641) Hypothetical GTP-binding protein yhbZ
Length = 390
Score = 137 bits (344), Expect = 9e-32
Identities = 83/220 (37%), Positives = 118/220 (52%), Gaps = 26/220 (11%)
Query: 239 IGEEQEEQEILYNVAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISN 298
+G +Q + ++T+ GQ+++VA+GG G GN + K
Sbjct: 92 VGTRVIDQGTGETMGDMTKHGQRLLVAKGGWHGLGNTRFKSSVNRTPRQKTN-------- 143
Query: 299 LEDSDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYA 358
G PG + L+LEL +ADV +GMPNAGKST + A+S AKP V Y
Sbjct: 144 -------------GTPGDKRELLLELMLLADVGMLGMPNAGKSTFIRAVSAAKPKVADYP 190
Query: 359 FTTLRPNLGNLNFD-DFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAA 417
FTTL P+LG + D + S VADIPGLI+GA + GLG FL+H+ER +VL +++D+
Sbjct: 191 FTTLVPSLGVVRMDNEKSFVVADIPGLIEGAAEGAGLGIRFLKHLERCRVLLHLIDIDPI 250
Query: 418 LHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANKID 457
G P E + +I ELE + L+ +P +V NKID
Sbjct: 251 ----DGTDPVENARIIISELEKYSQDLATKPRWLVFNKID 286
Score = 69.3 bits (168), Expect = 2e-11
Identities = 37/94 (39%), Positives = 55/94 (58%), Gaps = 4/94 (4%)
Query: 48 RMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPR---VWDFSG 104
+ +D+ I+ AGDGG+GC SF R ++ +G DGG+GG GGDV +E + D+
Sbjct: 2 KFVDEASILVVAGDGGNGCVSFRREKYIPKGGPDGGDGGDGGDVWMEADENLNTLIDYR- 60
Query: 105 LQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGT 138
+ A +G G+S++ G RG D + VPVGT
Sbjct: 61 FEKSFRAERGQNGASRDCTGKRGKDVTIKVPVGT 94
>Y384_MYCPN (P75215) Probable GTP-binding protein MG384 homolog
Length = 433
Score = 128 bits (321), Expect = 4e-29
Identities = 95/315 (30%), Positives = 143/315 (45%), Gaps = 30/315 (9%)
Query: 174 DHGNGSISNATNGEEVKTIHPTGCSSSQAAEKNVEKSVKSGHVASTDVLSQLSISNGTPK 233
D G N NG V C S + +SG D+ NG
Sbjct: 30 DKGGPGGGNGGNGGNVVLQADHNCDSLFFLKNKKHLFAESGGNGKPDLAHG---KNGEDL 86
Query: 234 FGTEYIGEEQEEQEILYNVAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACR 293
+G + + + + + Q I+ GG+GG GN + + PI
Sbjct: 87 VIKVPVGTTVRDLDTNQILMDFVHDQQSFILCYGGKGGKGNAAF----KSPI-------- 134
Query: 294 QHISNLEDSDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPA 353
NL ++ SL+ LE+K +A+V VG PN GKSTL+ +S AKP
Sbjct: 135 MRAPNLYENGDKGQSLHVS---------LEIKYLANVGIVGFPNTGKSTLISKLSNAKPK 185
Query: 354 VGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVD 413
+ +Y FTTL P LG + +D S+ ADIPGLI+ A + GLGH FLRHIER ++L +++
Sbjct: 186 IANYRFTTLVPVLGVVKHNDQSLVFADIPGLIENASEGSGLGHYFLRHIERCEILIHLIS 245
Query: 414 LAAALHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANK--IDEEGADEVYLELQRR 471
L H P + + ++ EL + L + L+VANK +D +G L
Sbjct: 246 LDPVDHD----DPCQAYEQIMRELSKYSQLLVKKKMLVVANKTDVDLDGTRFQKLAQYLE 301
Query: 472 VHGVPIFRVCAVLEE 486
G+P+F++ A+ +E
Sbjct: 302 NKGIPLFKISALKQE 316
Score = 60.8 bits (146), Expect = 8e-09
Identities = 33/84 (39%), Positives = 45/84 (53%), Gaps = 2/84 (2%)
Query: 59 AGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH--HLIAGKGGP 116
AG+GG+G ++ R H +G GGNGG GG+V+L+ L++ HL A GG
Sbjct: 13 AGNGGNGIIAWRREAHYDKGGPGGGNGGNGGNVVLQADHNCDSLFFLKNKKHLFAESGGN 72
Query: 117 GSSKNKIGTRGADKVVHVPVGTVI 140
G G G D V+ VPVGT +
Sbjct: 73 GKPDLAHGKNGEDLVIKVPVGTTV 96
>Y376_BUCAP (Q8K9G1) Hypothetical GTP-binding protein BUsg376
Length = 333
Score = 125 bits (313), Expect = 3e-28
Identities = 78/214 (36%), Positives = 117/214 (54%), Gaps = 27/214 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLS-TSKDSRKPITTKAGACRQHISNLEDSDSVCSSLN 310
+ +L + Q++++A+GG G GN S +R P RQ
Sbjct: 105 IGDLIKHKQKILIAKGGWHGLGNTRFKSSINRTP--------RQR--------------T 142
Query: 311 AGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLN 370
G G + + LEL IADV +GMPNAGKSTL+ +IS AK + +Y FTTL P LG++N
Sbjct: 143 LGSVGEKRDIQLELILIADVGTLGMPNAGKSTLVKSISGAKTKIANYPFTTLNPVLGSVN 202
Query: 371 FDDFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
+ +ADIPG+++ A Q GLG FL+H+ER K+L ++VDL H P E +
Sbjct: 203 TEGKKFIIADIPGIMQNASQGFGLGVRFLKHLERCKILLHIVDLCPTDHSN----PVENI 258
Query: 431 KDLILELEYHQDGLSNRPSLIVANKIDEEGADEV 464
+ ++ EL+ + L N+P ++ NKID + E+
Sbjct: 259 RIILNELKKYNTSLYNKPRWLILNKIDLIKSSEI 292
Score = 62.8 bits (151), Expect = 2e-09
Identities = 37/95 (38%), Positives = 54/95 (55%), Gaps = 2/95 (2%)
Query: 48 RMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH 107
+ ID+ I AG+GG+GC +F R ++ +G DGG+GG GG+V L+ + L+
Sbjct: 2 KFIDQTIIQVIAGNGGNGCVNFRREKYIPKGGPDGGDGGDGGNVWLQSDNNLNTLIDLRF 61
Query: 108 H--LIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
A G GS KN G +G+D ++VPVGT I
Sbjct: 62 KKTFQAPHGENGSGKNCSGKKGSDIKIYVPVGTKI 96
>Y384_MYCGE (P47624) Probable GTP-binding protein MG384
Length = 433
Score = 119 bits (299), Expect = 1e-26
Identities = 91/315 (28%), Positives = 143/315 (44%), Gaps = 30/315 (9%)
Query: 174 DHGNGSISNATNGEEVKTIHPTGCSSSQAAEKNVEKSVKSGHVASTDVLSQLSISNGTPK 233
D G N NG V C S + + G D+ NG+
Sbjct: 30 DKGGPGGGNGGNGGNVILQADHNCDSLFFLKNKKHLFAEDGQNGKPDLAHG---KNGSDL 86
Query: 234 FGTEYIGEEQEEQEILYNVAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACR 293
IG + E + + + Q I+ GG+GG GN + + PI
Sbjct: 87 LIKVPIGTTVKNLENNSVLVDFVHDKQSFILCFGGKGGKGNAAF----KSPI-------- 134
Query: 294 QHISNLEDSDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPA 353
NL ++ LN LE+K +A+V VG PN+GKSTL+ +S AKP
Sbjct: 135 MRAPNLYENGDKGEILNVS---------LEVKYLANVGIVGFPNSGKSTLISKLSNAKPK 185
Query: 354 VGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVD 413
+ +Y FTTL P LG + + + S+ ADIPGLI+ A + GLGH FLRHIER ++L +++
Sbjct: 186 IANYRFTTLIPVLGVVKYQNNSLVFADIPGLIENASEGSGLGHDFLRHIERCEILIHLIS 245
Query: 414 LAAALHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQR--R 471
L P + ++ EL + L + L+VANKID ++ + +L++ +
Sbjct: 246 LDPV----DNDDPCKAYLQIMDELSKYSPLLVKKKMLVVANKIDVNEGEKRFKKLEKFLQ 301
Query: 472 VHGVPIFRVCAVLEE 486
+ + ++ A+ +E
Sbjct: 302 KKSISVLKISALKKE 316
Score = 56.6 bits (135), Expect = 1e-07
Identities = 31/84 (36%), Positives = 45/84 (52%), Gaps = 2/84 (2%)
Query: 59 AGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH--HLIAGKGGP 116
AG+GG+G ++ R H +G GGNGG GG+VIL+ L++ HL A G
Sbjct: 13 AGNGGNGIIAWKREAHYDKGGPGGGNGGNGGNVILQADHNCDSLFFLKNKKHLFAEDGQN 72
Query: 117 GSSKNKIGTRGADKVVHVPVGTVI 140
G G G+D ++ VP+GT +
Sbjct: 73 GKPDLAHGKNGSDLLIKVPIGTTV 96
>Y389_BUCAI (P57469) Hypothetical GTP-binding protein BU389
Length = 334
Score = 118 bits (295), Expect = 4e-26
Identities = 75/215 (34%), Positives = 113/215 (51%), Gaps = 28/215 (13%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLS-TSKDSRKPITTKAGACRQHISNLEDSDSVCSSLN 310
+ +L + Q+M++A+GG G GN S +R P + G+
Sbjct: 105 IGDLIQHKQKMLIAKGGWHGLGNARFKSSTNRTPRQSTLGSI------------------ 146
Query: 311 AGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLN 370
G + + LEL +ADV +GMPN GKSTL+ IS AK + Y FTTL P LG++N
Sbjct: 147 ----GEKRDIQLELMLLADVGTLGMPNVGKSTLVTNISGAKTKISDYPFTTLHPVLGSVN 202
Query: 371 FD-DFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQ 429
+ +ADIPG+IKGA GLG FL+H+ER K+L +++DL + P +
Sbjct: 203 IQKNKKFIIADIPGIIKGASYGAGLGIRFLKHLERCKLLLHIIDLVP----QNNCHPSDN 258
Query: 430 LKDLILELEYHQDGLSNRPSLIVANKIDEEGADEV 464
+K ++ EL+ + L N+P + NKID +E+
Sbjct: 259 IKTVLNELKKYSLKLYNKPRWFIFNKIDLLSVEEL 293
Score = 67.4 bits (163), Expect = 8e-11
Identities = 37/95 (38%), Positives = 54/95 (55%), Gaps = 2/95 (2%)
Query: 48 RMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGL-- 105
+ ID+ I AG+GG+GC SF R ++ +G DGGNGG GG++ LE + + L
Sbjct: 2 KFIDQAIIHVIAGNGGNGCVSFRREKYIPKGGPDGGNGGDGGNIWLEANNNLNTLIDLRF 61
Query: 106 QHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
+ A G GSS+ G +G D +HVP+GT +
Sbjct: 62 KKKFQAQNGQNGSSRKSSGKKGDDIKIHVPIGTKV 96
>Y352_BUCBP (Q89AE7) Hypothetical GTP-binding protein bbp352
Length = 338
Score = 108 bits (271), Expect = 3e-23
Identities = 94/290 (32%), Positives = 143/290 (48%), Gaps = 47/290 (16%)
Query: 174 DHGNGS---ISNATNGEEVKTIHPTGCSSSQAAEKNVEKSVKSGHVASTDVLSQLSISNG 230
D GNG + TN + T +Q ++ K K+G S D++ Q+ I
Sbjct: 38 DGGNGGNVWLQTVTNLNTLIDFKFTKIFKAQDGQQGFNKK-KTGKKGS-DIVIQIPI--- 92
Query: 231 TPKFGTEYIGEEQEEQEILYNVAELTEEGQQMIVARGGEGGPGNLS-TSKDSRKPITTKA 289
GT+ I E + ++ ++ Q ++VA+GG G GN S +R PI
Sbjct: 93 ----GTKIIDHNTNEI-----IEDMIQDKQLVLVAKGGWHGLGNTRFKSSTNRIPI---- 139
Query: 290 GACRQHISNLEDSDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISR 349
+H G G +L LEL IA V +G+PN+GKSTL+ IS
Sbjct: 140 ----KHTK--------------GTQGEFRILRLELILIAHVGTLGLPNSGKSTLVRNISN 181
Query: 350 AKPAVGHYAFTTLRPNLG--NLNFDDFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKV 407
AK + +Y FTTL+P LG +N +F + +ADIPGLI+GA GLG+ FL+H+ER +
Sbjct: 182 AKTKIANYPFTTLKPVLGTVKINHKEFFV-IADIPGLIQGASHGIGLGYQFLKHLERCHL 240
Query: 408 LAYVVDLAAALHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANKID 457
L +++D+ + ++ + I + D EL+ + L N+P V NKID
Sbjct: 241 LLHIIDI-SQINFKNTITNIHVILD---ELKTYNKILHNKPIWFVFNKID 286
Score = 61.6 bits (148), Expect = 5e-09
Identities = 38/109 (34%), Positives = 59/109 (53%), Gaps = 4/109 (3%)
Query: 48 RMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH 107
+ +DK I AG+GG G +SF R ++ +G DGG+GG GG+V L+ + +
Sbjct: 2 KFLDKAIIHVIAGNGGHGRTSFRREKYIPKGGPDGGDGGNGGNVWLQTVTNLNTLIDFKF 61
Query: 108 HLI--AGKGGPGSSKNKIGTRGADKVVHVPVGTVI--HLVNGDIPSIVK 152
I A G G +K K G +G+D V+ +P+GT I H N I +++
Sbjct: 62 TKIFKAQDGQQGFNKKKTGKKGSDIVIQIPIGTKIIDHNTNEIIEDMIQ 110
>GTP1_SCHPO (P32235) GTP-binding protein 1
Length = 364
Score = 68.9 bits (167), Expect = 3e-11
Identities = 39/97 (40%), Positives = 54/97 (55%), Gaps = 1/97 (1%)
Query: 328 ADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKG 387
A V+F+G P+ GKSTLL AI++ K A Y FTTL G L +D I + D+PG+I+G
Sbjct: 63 ARVAFIGFPSVGKSTLLSAITKTKSATASYEFTTLTAIPGVLEYDGAEIQMLDLPGIIEG 122
Query: 388 AHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGI 424
A Q RG G + ++ V+D A R+ I
Sbjct: 123 ASQGRG-GRQAVSAARTADLILMVLDATKAADQREKI 158
>YG3Y_YEAST (P53295) Hypothetical 41.0 kDa protein in YIP1-CBP4
intergenic region
Length = 368
Score = 68.6 bits (166), Expect = 4e-11
Identities = 35/97 (36%), Positives = 51/97 (52%)
Query: 328 ADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKG 387
A V +G P+ GKS+LLG I+ K + HYAFTTL G L + I + D+PG+I G
Sbjct: 64 ARVVLIGYPSVGKSSLLGKITTTKSEIAHYAFTTLTSVPGVLKYQGAEIQIVDLPGIIYG 123
Query: 388 AHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGI 424
A Q +G G + ++ V+D + H R +
Sbjct: 124 ASQGKGRGRQVVATARTADLVLMVLDATKSEHQRASL 160
>YD26_METJA (Q58722) Hypothetical GTP-binding protein MJ1326
Length = 391
Score = 66.2 bits (160), Expect = 2e-10
Identities = 56/185 (30%), Positives = 86/185 (46%), Gaps = 22/185 (11%)
Query: 328 ADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKG 387
A +FVG P+ GKSTLL ++ AK VG YAFTTL G L + I + D PG+I G
Sbjct: 85 ATAAFVGFPSVGKSTLLNKLTNAKSEVGAYAFTTLTIVPGILEYKGAKIQLLDAPGIIVG 144
Query: 388 AHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQLKDLI------------L 435
A +G G L + ++ VD+ H +P E K+L +
Sbjct: 145 ASSGKGRGTEVLSAVRSADLILLTVDIYTLDH----LPVLE--KELYNVGIRLDQTPPDV 198
Query: 436 ELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVP-IFRVCAVLEEGIPELKAG 494
+++ + G N S + IDE+ + + E R+H + R LE+ I ++ AG
Sbjct: 199 KIKVKERGGINVSSTVPLTHIDEDTIEAILNEY--RIHNADVVIREDITLEQFI-DVVAG 255
Query: 495 LRMLV 499
R+ +
Sbjct: 256 NRVYI 260
>YB11_HALN1 (P17103) Hypothetical protein Vng1111g
Length = 370
Score = 65.5 bits (158), Expect = 3e-10
Identities = 35/85 (41%), Positives = 50/85 (58%)
Query: 328 ADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKG 387
A V+ VG P+ GKS+L+ A++ A VG Y FTTL N G L + +I + D+PGLI+G
Sbjct: 62 ATVALVGFPSVGKSSLINAMTNADSEVGAYEFTTLNVNPGMLEYRGANIQLLDVPGLIEG 121
Query: 388 AHQNRGLGHAFLRHIERTKVLAYVV 412
A RG G L I ++ +V+
Sbjct: 122 AAGGRGGGKEILSVIRGADLVIFVL 146
>Y024_MYCGE (P47270) Probable GTP-binding protein MG024
Length = 367
Score = 65.5 bits (158), Expect = 3e-10
Identities = 35/97 (36%), Positives = 50/97 (51%), Gaps = 17/97 (17%)
Query: 333 VGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDD-----------------FS 375
VG+PN GKSTL AI+ + + +Y F T+ PN G +N D +
Sbjct: 7 VGLPNVGKSTLFSAITNLQVEIANYPFATIEPNTGIVNVSDERLDKLASLINPEKIVYTT 66
Query: 376 ITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVV 412
DI GL+KGA Q +GLG+ FL +I ++ +VV
Sbjct: 67 FRFVDIAGLVKGASQGQGLGNQFLANIREVDLICHVV 103
>ENGD_BUCAP (Q8K9V2) GTP-dependent nucleic acid-binding protein engD
Length = 361
Score = 65.1 bits (157), Expect = 4e-10
Identities = 34/97 (35%), Positives = 52/97 (53%), Gaps = 17/97 (17%)
Query: 333 VGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSIT--------------- 377
+G+PN GKSTL +++ AV ++ F T++PN+G ++ D I
Sbjct: 7 IGLPNVGKSTLFNVLTKGNSAVANFPFCTIKPNIGIVSVPDNRINNLSKIILPKKITNAF 66
Query: 378 --VADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVV 412
DI GL+KGA + GLG+ FL +I T +A+VV
Sbjct: 67 IEFVDIAGLVKGASKGEGLGNQFLSNIRDTHAIAHVV 103
>ENGD_BACSU (P37518) GTP-dependent nucleic acid-binding protein engD
Length = 366
Score = 62.8 bits (151), Expect = 2e-09
Identities = 39/125 (31%), Positives = 60/125 (47%), Gaps = 20/125 (16%)
Query: 333 VGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDD-----------------FS 375
VG+PN GKSTL AI++A +Y F T+ PN+G + D +
Sbjct: 8 VGLPNVGKSTLFNAITQAGAESANYPFCTIDPNVGIVEVPDDRLQKLTELVNPKKTVPTA 67
Query: 376 ITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAA---ALHGRKGIPPWEQLKD 432
DI G++KGA + GLG+ FL HI + + +VV + H + P + ++
Sbjct: 68 FEFTDIAGIVKGASKGEGLGNKFLSHIRQVDAICHVVRAFSDDNITHVSGKVDPIDDIET 127
Query: 433 LILEL 437
+ LEL
Sbjct: 128 INLEL 132
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 63,680,992
Number of Sequences: 164201
Number of extensions: 2999292
Number of successful extensions: 7921
Number of sequences better than 10.0: 314
Number of HSP's better than 10.0 without gapping: 79
Number of HSP's successfully gapped in prelim test: 237
Number of HSP's that attempted gapping in prelim test: 7581
Number of HSP's gapped (non-prelim): 486
length of query: 515
length of database: 59,974,054
effective HSP length: 115
effective length of query: 400
effective length of database: 41,090,939
effective search space: 16436375600
effective search space used: 16436375600
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0176a.13


