

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0175.13
(437 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
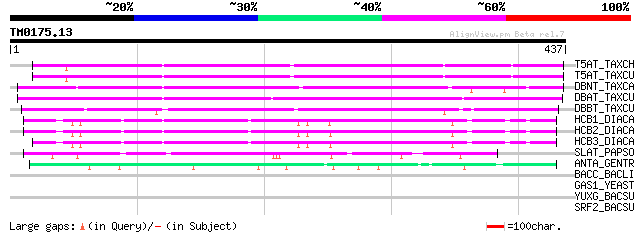
Score E
Sequences producing significant alignments: (bits) Value
T5AT_TAXCH (Q8S9G6) Taxadien-5-alpha-ol O-acetyltransferase (EC ... 233 7e-61
T5AT_TAXCU (Q9M6F0) Taxadien-5-alpha-ol O-acetyltransferase (EC ... 231 3e-60
DBNT_TAXCA (Q8LL69) 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransf... 223 1e-57
DBAT_TAXCU (Q9M6E2) 10-deacetylbaccatin III 10-O-acetyltransfera... 216 1e-55
DBBT_TAXCU (Q9FPW3) 2-alpha-hydroxytaxane 2-O-benzoyltransferase... 207 6e-53
HCB1_DIACA (O24645) Anthranilate N-benzoyltransferase protein 1 ... 152 1e-36
HCB2_DIACA (O23917) Anthranilate N-benzoyltransferase protein 2 ... 147 4e-35
HCB3_DIACA (O23918) Anthranilate N-benzoyltransferase protein 3 ... 146 9e-35
SLAT_PAPSO (Q94FT4) Salutaridinol 7-O-acetyltransferase (EC 2.3.... 96 2e-19
ANTA_GENTR (Q9ZWR8) Anthocyanin 5-aromatic acyltransferase (EC 2... 59 2e-08
BACC_BACLI (O68008) Bacitracin synthetase 3 (BA3) [Includes: ATP... 39 0.026
GAS1_YEAST (P22146) Glycolipid anchored surface protein 1 precur... 31 5.5
YUXG_BACSU (P40747) Hypothetical oxidoreductase yuxG (EC 1.-.-.-... 31 7.2
SRF2_BACSU (Q04747) Surfactin synthetase subunit 2 31 7.2
>T5AT_TAXCH (Q8S9G6) Taxadien-5-alpha-ol O-acetyltransferase (EC
2.3.1.162)
(Taxa-4(20),11(12)-dien-5alpha-ol-O-acetyltransferase)
(Taxadienol acetyltransferase)
Length = 439
Score = 233 bits (594), Expect = 7e-61
Identities = 146/423 (34%), Positives = 217/423 (50%), Gaps = 10/423 (2%)
Query: 19 LVPPASSTPHEVKLLSDIDDQDGLR---FNIPFIMMYRHEPSMADKDPVQAIRQALSRTL 75
+V P+ P LS ID+ G+R FN I P+M DP + IR+AL++ L
Sbjct: 15 MVGPSLPLPKTTLQLSSIDNLPGVRGSIFNALLIYNASPSPTMVSADPAKLIREALAKIL 74
Query: 76 VYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVP 135
VYY PFAGRL+E L V+CTGEG MF+EA AD L G+ P F++LL+ +P
Sbjct: 75 VYYPPFAGRLRETENGDLEVECTGEGAMFLEAMADNELSVLGDFD-DSNPSFQQLLFSLP 133
Query: 136 GSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTPP 195
D PLL++QVTR CGGF++ + F+H + DG G QFL LAEMA+G + P
Sbjct: 134 LDTNFKDLPLLVVQVTRFTCGGFVVGVSFHHGVCDGRGAAQFLKGLAEMARGEVKLSLEP 193
Query: 196 VWCRELLMARDPPRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRRLVPLDLRH- 254
+W REL+ DP + H+EF + P S E + F I +++ V + +
Sbjct: 194 IWNRELVKLDDPKYLQFFHFEFLRAP--SIVEKIVQTYFIIDFETINYIKQSVMEECKEF 251
Query: 255 CSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTT 314
CS+F+V +A W RT+A Q+ + ++++ ++ RN FNPP+P GYYGN V
Sbjct: 252 CSSFEVASAMTWIARTRAFQIPESEYVKILFGMDMRNSFNPPLPSGYYGNSIGTACAVDN 311
Query: 315 VGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRSCIVSDLTRFKLH 374
V L S A+ +++K+K + + S A + +D +R
Sbjct: 312 VQDLLSGSLLRAIMIIKKSKVSLNDNF-KSRAVVKPSELDVNMNHENVVAFADWSRLGFD 370
Query: 375 ETDFGWGEPVCGGVAKGGAGLYGGASYIIACKNAKGE-DGRVLVICLPVENMKRFAKELN 433
E DFGWG V + L +Y + K +K + DG +++ LP+ MK F E+
Sbjct: 371 EVDFGWGNAVSVSPVQQQCEL-AMQNYFLFLKPSKNKPDGIKILMFLPLSKMKSFKIEME 429
Query: 434 NMI 436
M+
Sbjct: 430 AMM 432
>T5AT_TAXCU (Q9M6F0) Taxadien-5-alpha-ol O-acetyltransferase (EC
2.3.1.162)
(Taxa-4(20),11(12)-dien-5alpha-ol-O-acetyltransferase)
(Taxadienol acetyltransferase)
Length = 439
Score = 231 bits (589), Expect = 3e-60
Identities = 145/423 (34%), Positives = 217/423 (51%), Gaps = 10/423 (2%)
Query: 19 LVPPASSTPHEVKLLSDIDDQDGLR---FNIPFIMMYRHEPSMADKDPVQAIRQALSRTL 75
+V P+ P LS ID+ G+R FN I P+M DP + IR+AL++ L
Sbjct: 15 MVGPSPPLPKTTLQLSSIDNLPGVRGSIFNALLIYNASPSPTMISADPAKPIREALAKIL 74
Query: 76 VYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVP 135
VYY PFAGRL+E L V+CTGEG MF+EA AD L G+ P F++LL+ +P
Sbjct: 75 VYYPPFAGRLRETENGDLEVECTGEGAMFLEAMADNELSVLGDFD-DSNPSFQQLLFSLP 133
Query: 136 GSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTPP 195
D LL++QVTR CGGF++ + F+H + DG G QFL LAEMA+G + P
Sbjct: 134 LDTNFKDLSLLVVQVTRFTCGGFVVGVSFHHGVCDGRGAAQFLKGLAEMARGEVKLSLEP 193
Query: 196 VWCRELLMARDPPRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRRLVPLDLRH- 254
+W REL+ DP + H+EF + P S E + F I +++ V + +
Sbjct: 194 IWNRELVKLDDPKYLQFFHFEFLRAP--SIVEKIVQTYFIIDFETINYIKQSVMEECKEF 251
Query: 255 CSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTT 314
CS+F+V +A W RT+A Q+ + ++++ ++ RN FNPP+P GYYGN V
Sbjct: 252 CSSFEVASAMTWIARTRAFQIPESEYVKILFGMDMRNSFNPPLPSGYYGNSIGTACAVDN 311
Query: 315 VGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRSCIVSDLTRFKLH 374
V L S A+ +++K+K + + S A + +D +R
Sbjct: 312 VQDLLSGSLLRAIMIIKKSKVSLNDNF-KSRAVVKPSELDVNMNHENVVAFADWSRLGFD 370
Query: 375 ETDFGWGEPVCGGVAKGGAGLYGGASYIIACKNAKGE-DGRVLVICLPVENMKRFAKELN 433
E DFGWG V + + L +Y + K +K + DG +++ LP+ MK F E+
Sbjct: 371 EVDFGWGNAVSVSPVQQQSAL-AMQNYFLFLKPSKNKPDGIKILMFLPLSKMKSFKIEME 429
Query: 434 NMI 436
M+
Sbjct: 430 AMM 432
>DBNT_TAXCA (Q8LL69) 3'-N-debenzoyl-2'-deoxytaxol
N-benzoyltransferase (EC 2.3.1.-) (DBTNBT)
Length = 441
Score = 223 bits (567), Expect = 1e-57
Identities = 141/437 (32%), Positives = 226/437 (51%), Gaps = 15/437 (3%)
Query: 7 SLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQA 66
S F V++ P +V P+ +P LS +D R ++++ + P DPV+
Sbjct: 6 STDFHVKKFDPVMVAPSLPSPKATVQLSVVDSLTICRGIFNTLLVF-NAPDNISADPVKI 64
Query: 67 IRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPC 126
IR+ALS+ LVYY+P AGRL+ +L V+CTG+G +F+EA + ++ + P
Sbjct: 65 IREALSKVLVYYFPLAGRLRSKEIGELEVECTGDGALFVEAMVEDTISVLRDLD-DLNPS 123
Query: 127 FEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQ 186
F++L++ P + D L+++QVTR CGG + + H++ DG G QF++ALAEMA+
Sbjct: 124 FQQLVFWHPLDTAIEDLHLVIVQVTRFTCGGIAVGVTLPHSVCDGRGAAQFVTALAEMAR 183
Query: 187 GASQPLTPPVWCRELLMARDPPRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRR 246
G +P P+W RELL DP + N ++ P E G SF + I +++
Sbjct: 184 GEVKPSLEPIWNRELLNPEDPLHLQLNQFDSICPPPMLEELG--QASFVINVDTIEYMKQ 241
Query: 247 LVPLDLRH-CSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNC 305
V + CS+F+V+ A W RTKALQ+ ++++L+ ++ R FNPP+P GYYGN
Sbjct: 242 CVMEECNEFCSSFEVVAALVWIARTKALQIPHTENVKLLFAMDLRKLFNPPLPNGYYGNA 301
Query: 306 FTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRS--- 362
+ V L S A+ +++KAKA + Y S +V N L +S
Sbjct: 302 IGTAYAMDNVQDLLNGSLLRAIMIIKKAKADLKDNYSRSR---VVTNPYSLDVNKKSDNI 358
Query: 363 CIVSDLTRFKLHETDFGWGEPVCGGV---AKGGAGLYGGASYIIACKNAKGEDGRVLVIC 419
+SD R +E DFGWG P+ + G ++ Y++ KN K + ++L+ C
Sbjct: 359 LALSDWRRLGFYEADFGWGGPLNVSSLQRLENGLPMFSTFLYLLPAKN-KSDGIKLLLSC 417
Query: 420 LPVENMKRFAKELNNMI 436
+P +K F + MI
Sbjct: 418 MPPTTLKSFKIVMEAMI 434
>DBAT_TAXCU (Q9M6E2) 10-deacetylbaccatin III 10-O-acetyltransferase
(EC 2.3.1.167) (DBAT)
Length = 440
Score = 216 bits (549), Expect = 1e-55
Identities = 138/432 (31%), Positives = 214/432 (48%), Gaps = 6/432 (1%)
Query: 7 SLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQA 66
S F V+ + +V P+ +P LS +D+ G+R NI ++ + DP +
Sbjct: 4 STEFVVRSLERVMVAPSQPSPKAFLQLSTLDNLPGVRENIFNTLLVYNASDRVSVDPAKV 63
Query: 67 IRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPC 126
IRQALS+ LVYY PFAGRL++ L V+CTGEG +F+EA AD L G+ P
Sbjct: 64 IRQALSKVLVYYSPFAGRLRKKENGDLEVECTGEGALFVEAMADTDLSVLGDLD-DYSPS 122
Query: 127 FEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQ 186
E+LL+ +P + D L++QVTR CGGF++ + F H + DG G QFL A+ EMA+
Sbjct: 123 LEQLLFCLPPDTDIEDIHPLVVQVTRFTCGGFVVGVSFCHGICDGLGAGQFLIAMGEMAR 182
Query: 187 GASQPLTPPVWCRELLMARDPPRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRR 246
G +P + P+W RELL D P +Y F+ + ST + S S I +++
Sbjct: 183 GEIKPSSEPIWKRELLKPED-PLYRFQYYHFQLICPPSTFGKIVQGSLVITSETINCIKQ 241
Query: 247 LVPLDLRH-CSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNC 305
+ + + CS F+V++A W RT+ALQ+ ++++L+ ++ R FNPP+ GYYGN
Sbjct: 242 CLREESKEFCSAFEVVSALAWIARTRALQIPHSENVKLIFAMDMRKLFNPPLSKGYYGNF 301
Query: 306 FTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRSCIV 365
+ V L S V +++KAK E + ++ + +
Sbjct: 302 VGTVCAMDNVKDLLSGSLLRVVRIIKKAKVSLNEHFTSTIVTPRSGSDESI-NYENIVGF 360
Query: 366 SDLTRFKLHETDFGWGEPVCGGVAKGG-AGLYGGASYIIACKNAKGE-DGRVLVICLPVE 423
D R E DFGWG + + G + SY + + K DG ++ +P
Sbjct: 361 GDRRRLGFDEVDFGWGHADNVSLVQHGLKDVSVVQSYFLFIRPPKNNPDGIKILSFMPPS 420
Query: 424 NMKRFAKELNNM 435
+K F E+ M
Sbjct: 421 IVKSFKFEMETM 432
>DBBT_TAXCU (Q9FPW3) 2-alpha-hydroxytaxane 2-O-benzoyltransferase
(EC 2.3.1.166) (TBT) (2-debenzoyl-7,13-diacetylbaccatin
III-2-O-benzoyl transferase) (DBBT)
Length = 440
Score = 207 bits (526), Expect = 6e-53
Identities = 135/432 (31%), Positives = 220/432 (50%), Gaps = 20/432 (4%)
Query: 10 FKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQ 69
F V + +V P +P + LS ID++ NI + S++ DP + IR+
Sbjct: 4 FNVDMIERVIVAPCLQSPKNILHLSPIDNKTRGLTNILSVYNASQRVSVS-ADPAKTIRE 62
Query: 70 ALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLV---EFGETPLPPFPC 126
ALS+ LVYY PFAGRL+ L V+CTGEG +F+EA AD L +F E P
Sbjct: 63 ALSKVLVYYPPFAGRLRNTENGDLEVECTGEGAVFVEAMADNDLSVLQDFNEYD----PS 118
Query: 127 FEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQ 186
F++L++++ + D LL +QVTR CGGF++ RF+H++SDG G+ Q L + EMA+
Sbjct: 119 FQQLVFNLREDVNIEDLHLLTVQVTRFTCGGFVVGTRFHHSVSDGKGIGQLLKGMGEMAR 178
Query: 187 GASQPLTPPVWCRELLMARDPPRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRR 246
G +P P+W RE++ D + +H++F P + E +I S I ++R
Sbjct: 179 GEFKPSLEPIWNREMVKPEDIMYLQFDHFDFIHPPLNL--EKSIQASMVISFERINYIKR 236
Query: 247 LVPLDLRH-CSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNC 305
+ + + S F+V+ A W RTK+ ++ P++ ++++ ++ RN F+ P+P GYYGN
Sbjct: 237 CMMEECKEFFSAFEVVVALIWLARTKSFRIPPNEYVKIIFPIDMRNSFDSPLPKGYYGNA 296
Query: 306 FTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEY---MHSMADFLVANRRCLFTTVRS 362
+ V L S YA+ L++K+K E + + + L AN + V
Sbjct: 297 IGNACAMDNVKDLLNGSLLYALMLIKKSKFALNENFKSRILTKPSTLDANMK--HENVVG 354
Query: 363 CIVSDLTRFKLHETDFGWGEPV-CGGVAKGGAGLYGGASYIIACKNAKGE-DGRVLVICL 420
C D +E DFGWG V + + +Y + ++AK DG +++ +
Sbjct: 355 C--GDWRNLGFYEADFGWGNAVNVSPMQQQREHELAMQNYFLFLRSAKNMIDGIKILMFM 412
Query: 421 PVENMKRFAKEL 432
P +K F E+
Sbjct: 413 PASMVKPFKIEM 424
>HCB1_DIACA (O24645) Anthranilate N-benzoyltransferase protein 1 (EC
2.3.1.144) (Anthranilate
N-hydroxycinnamoyl/benzoyltransferase 1)
Length = 445
Score = 152 bits (385), Expect = 1e-36
Identities = 130/453 (28%), Positives = 214/453 (46%), Gaps = 49/453 (10%)
Query: 12 VQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPF-----IMMYRH--------EPSM 58
+Q + +V PA TP++ LS+ID + P+ +++Y+ PS
Sbjct: 3 IQIKQSTMVRPAEETPNKSLWLSNID----MILRTPYSHTGAVLIYKQPDNNEDNIHPSS 58
Query: 59 ADKDPVQAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGE 118
+ + +ALS+ LV +YP AGRLK G + +DC EG +F+EA + L +FG+
Sbjct: 59 SMYFDANILIEALSKALVPFYPMAGRLKIN-GDRYEIDCNAEGALFVEAESSHVLEDFGD 117
Query: 119 TPLPPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFL 178
P ++ S+ + PLL++Q+TR +CGG + +H + DG +F
Sbjct: 118 FR-PNDELHRVMVPTCDYSKGISSFPLLMVQLTRFRCGGVSIGFAQHHHVCDGMAHFEFN 176
Query: 179 SALAEMAQGASQPLTPPVWCREL-LMARDPPRITCNHYEFEQ-VPSDSTE--EGAITRS- 233
++ A +A+G P PV R L L R+PP+I +H +FE VPS E +G +S
Sbjct: 177 NSWARIAKGL-LPALEPVHDRYLHLRPRNPPQIKYSHSQFEPFVPSLPNELLDGKTNKSQ 235
Query: 234 --FFFGSNEIAALRRLVPL--DLRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNT 289
F +I L++ + L + ST++V+ A W +KA L+ H++I+L+ V+
Sbjct: 236 TLFILSREQINTLKQKLDLSNNTTRLSTYEVVAAHVWRSVSKARGLSDHEEIKLIMPVDG 295
Query: 290 RNRF-NPPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMAD- 347
R+R NP +P GY GN TVG L N V++A ++Y+ S D
Sbjct: 296 RSRINNPSLPKGYCGNVVFLAVCTATVGDLSCNPLTDTAGKVQEALKGLDDDYLRSAIDH 355
Query: 348 ----------FLVANRRCLFTTVRSCIVSDLTRFKLHETDFGWGEPVCGGVAKGGAGLYG 397
++ + + L+ V +V+ R DFGWG P G++ Y
Sbjct: 356 TESKPGLPVPYMGSPEKTLYPNV---LVNSWGRIPYQAMDFGWGSPTFFGISN---IFYD 409
Query: 398 GASYIIACKNAKGEDGRVLVICLPVENMKRFAK 430
G ++I ++ G+ L I L ++ RF K
Sbjct: 410 GQCFLIPSRD--GDGSMTLAINLFSSHLSRFKK 440
>HCB2_DIACA (O23917) Anthranilate N-benzoyltransferase protein 2 (EC
2.3.1.144) (Anthranilate
N-hydroxycinnamoyl/benzoyltransferase 2)
Length = 446
Score = 147 bits (372), Expect = 4e-35
Identities = 130/454 (28%), Positives = 208/454 (45%), Gaps = 50/454 (11%)
Query: 12 VQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPF-----IMMYRH--------EPSM 58
+Q + +V PA TP++ LS ID + P+ +++Y+ PS
Sbjct: 3 IQIKQSTMVRPAEETPNKSLWLSKID----MILRTPYSHTGAVLIYKQPDNNEDNIHPSS 58
Query: 59 ADKDPVQAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGE 118
+ + +ALS+ LV YYP AGRLK G + +DC EG +F+EA + L +FG+
Sbjct: 59 SMYFDANILIEALSKALVPYYPMAGRLKIN-GDRYEIDCNAEGALFVEAESSHVLEDFGD 117
Query: 119 TPLPPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFL 178
P ++ S+ + PLL++Q+TR +CGG + +H DG +F
Sbjct: 118 FR-PNDELHRVMVPTCDYSKGISSFPLLMVQLTRFRCGGVSIGFAQHHHACDGMSHFEFN 176
Query: 179 SALAEMAQGASQPLTPPVWCREL-LMARDPPRITCNHYEFEQ-VPSDSTE--EGAITRS- 233
++ A +A+G P PV R L L R+PP+I H +FE VPS E +G +S
Sbjct: 177 NSWARIAKGL-LPALEPVHDRYLHLRLRNPPQIKYTHSQFEPFVPSLPNELLDGKTNKSQ 235
Query: 234 --FFFGSNEIAALRRLVPLD---LRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVN 288
F +I L++ + L ST++V+ W +KA L+ H++I+L+ V+
Sbjct: 236 TLFKLSREQINTLKQKLDLSSNTTTRLSTYEVVAGHVWRSVSKARGLSDHEEIKLIMPVD 295
Query: 289 TRNRF-NPPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMAD 347
R+R NP +P GY GN TVG L N V++A ++Y+ S D
Sbjct: 296 GRSRINNPSLPKGYCGNVVFLAVCTATVGDLSCNPLTDTAGKVQEALKGLDDDYLRSAID 355
Query: 348 -----------FLVANRRCLFTTVRSCIVSDLTRFKLHETDFGWGEPVCGGVAKGGAGLY 396
++ + + L+ V +V+ R DFGWG P G++ Y
Sbjct: 356 HTESKPDLPVPYMGSPEKTLYPNV---LVNSWGRIPYQAMDFGWGSPTFFGISN---IFY 409
Query: 397 GGASYIIACKNAKGEDGRVLVICLPVENMKRFAK 430
G ++I +N G+ L I L ++ F K
Sbjct: 410 DGQCFLIPSQN--GDGSMTLAINLFSSHLSLFKK 441
>HCB3_DIACA (O23918) Anthranilate N-benzoyltransferase protein 3 (EC
2.3.1.144) (Anthranilate
N-hydroxycinnamoyl/benzoyltransferase 3)
Length = 445
Score = 146 bits (369), Expect = 9e-35
Identities = 128/446 (28%), Positives = 207/446 (45%), Gaps = 49/446 (10%)
Query: 19 LVPPASSTPHEVKLLSDIDDQDGLRFNIPF-----IMMYRH--------EPSMADKDPVQ 65
+V PA TP++ LS ID + P+ +++Y+ +PS +
Sbjct: 10 MVRPAEETPNKSLWLSKID----MILRTPYSHTGAVLIYKQPDNNEDNIQPSSSMYFDAN 65
Query: 66 AIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFP 125
+ +ALS+ LV YYP AGRLK G + +DC GEG +F+EA + L +FG+ P
Sbjct: 66 ILIEALSKALVPYYPMAGRLKIN-GDRYEIDCNGEGALFVEAESSHVLEDFGDFR-PNDE 123
Query: 126 CFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMA 185
++ S+ + PLL++Q+TR +CGG + +H + D +F ++ A +A
Sbjct: 124 LHRVMVPTCDYSKGISSFPLLMVQLTRFRCGGVSIGFAQHHHVCDRMSHFEFNNSWARIA 183
Query: 186 QGASQPLTPPVWCREL-LMARDPPRITCNHYEFEQ-VPSDSTE--EGAITRS---FFFGS 238
+G P PV R L L R+PP+I H +FE VPS E +G ++S F
Sbjct: 184 KGL-LPALEPVHDRYLHLCPRNPPQIKYTHSQFEPFVPSLPKELLDGKTSKSQTLFKLSR 242
Query: 239 NEIAALRRLVPLD--LRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRF-NP 295
+I L++ + ST++V+ W +KA L+ H++I+L+ V+ R+R NP
Sbjct: 243 EQINTLKQKLDWSNTTTRLSTYEVVAGHVWRSVSKARGLSDHEEIKLIMPVDGRSRINNP 302
Query: 296 PIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMAD-------- 347
+P GY GN TVG L N V++A ++Y+ S D
Sbjct: 303 SLPKGYCGNVVFLAVCTATVGDLACNPLTDTAGKVQEALKGLDDDYLRSAIDHTESKPDL 362
Query: 348 ---FLVANRRCLFTTVRSCIVSDLTRFKLHETDFGWGEPVCGGVAKGGAGLYGGASYIIA 404
++ + + L+ V +V+ R DFGWG P G++ Y G ++I
Sbjct: 363 PVPYMGSPEKTLYPNV---LVNSWGRIPYQAMDFGWGNPTFFGISN---IFYDGQCFLIP 416
Query: 405 CKNAKGEDGRVLVICLPVENMKRFAK 430
+N G+ L I L ++ F K
Sbjct: 417 SQN--GDGSMTLAINLFSSHLSLFKK 440
>SLAT_PAPSO (Q94FT4) Salutaridinol 7-O-acetyltransferase (EC
2.3.1.150) (salAT)
Length = 474
Score = 95.5 bits (236), Expect = 2e-19
Identities = 98/433 (22%), Positives = 178/433 (40%), Gaps = 78/433 (18%)
Query: 12 VQRCKPELVPPASSTPHEVKL--LSDIDDQDGLRFNIPFIMMY-----RHEPSMADKDPV 64
V+ E + P + TP ++K LS +D L + +P I+ Y S D +
Sbjct: 9 VEVISKETIKPTTPTPSQLKNFNLSLLDQCFPLYYYVPIILFYPATAANSTGSSNHHDDL 68
Query: 65 QAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPF 124
++ +LS+TLV++YP AGR+ + ++VDC +G+ F + + EF P P
Sbjct: 69 DLLKSSLSKTLVHFYPMAGRMID----NILVDCHDQGINFYKVKIRGKMCEFMSQPDVP- 123
Query: 125 PCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEM 184
+LL S V L+++QV CGG + +H ++D A + F+ + A
Sbjct: 124 --LSQLLPSEVVSASVPKEALVIVQVNMFDCGGTAICSSVSHKIADAATMSTFIRSWAST 181
Query: 185 AQ-GASQPLTPPVWCRELLMARD-----PP--RIT----CNHYEFEQVPSDSTEEGAITR 232
+ S T V ++L+ + D PP R+T + F P D+ ++ +++
Sbjct: 182 TKTSRSGGSTAAVTDQKLIPSFDSASLFPPSERLTSPSGMSEIPFSSTPEDTEDDKTVSK 241
Query: 233 SFFFGSNEIAALRRLVPLDL------RHCSTFDVITACFWYCRTKALQLAPHDDIRLM-T 285
F F +I ++R + + + R + +V+T+ W ++ P + ++
Sbjct: 242 RFVFDFAKITSVREKLQVLMHDNYKSRRQTRVEVVTSLIW---KSVMKSTPAGFLPVVHH 298
Query: 286 IVNTRNRFNPPIPVGYYGNCFT-----YPAVVTTVGKLCGNSFGYAVELVRKAKAQATEE 340
VN R + +PP+ +GN PA TT AV + + ++
Sbjct: 299 AVNLRKKMDPPLQDVSFGNLSVTVSAFLPATTTTTTN--------AVNKTINSTSSESQV 350
Query: 341 YMHSMADFLVANR-----------------------------RCLFTTVRSCIVSDLTRF 371
+H + DF+ R + V + +S R
Sbjct: 351 VLHELHDFIAQMRSEIDKVKGDKGSLEKVIQNFASGHDASIKKINDVEVINFWISSWCRM 410
Query: 372 KLHETDFGWGEPV 384
L+E DFGWG+P+
Sbjct: 411 GLYEIDFGWGKPI 423
>ANTA_GENTR (Q9ZWR8) Anthocyanin 5-aromatic acyltransferase (EC
2.3.1.153) (5AT)
Length = 469
Score = 58.9 bits (141), Expect = 2e-08
Identities = 104/457 (22%), Positives = 172/457 (36%), Gaps = 51/457 (11%)
Query: 16 KPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADK---DPVQAIRQALS 72
K ++ PP+ +T E+ L D L N +++ P + ++ +LS
Sbjct: 12 KCQVTPPSDTTDVELSLPVTFFDIPWLHLNKMQSLLFYDFPYPRTHFLDTVIPNLKASLS 71
Query: 73 RTLVYYYPFAGRL----KEGPGRKLMVDCT-GEGVMFIEANADVSLVEFGETPLPPFPCF 127
TL +Y P +G L K G K G+ + I A +D L
Sbjct: 72 LTLKHYVPLSGNLLMPIKSGEMPKFQYSRDEGDSITLIVAESDQDFDYLKGHQLVDSNDL 131
Query: 128 EELLYDVPGSEQVLDS----PLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAE 183
L Y +P + + PL+ +QVT G +AL +H+++D F++A A
Sbjct: 132 HGLFYVMPRVIRTMQDYKVIPLVAVQVTVFPNRGIAVALTAHHSIADAKSFVMFINAWAY 191
Query: 184 MAQ-GASQPLTP----PVWCRELLMARDPPRITC-NHYE-----FEQVPSDSTEEGAITR 232
+ + G L P + R ++ T N + F + S +
Sbjct: 192 INKFGKDADLLSANLLPSFDRSIIKDLYGLEETFWNEMQDVLEMFSRFGSKPPRFNKVRA 251
Query: 233 SFFFGSNEIAALRRLVPLDLR------HCSTFDVITACFWYCRTKAL-----QLAPHDDI 281
++ EI L+ V L+LR +TF + W C K+ + + +D+
Sbjct: 252 TYVLSLAEIQKLKNKV-LNLRGSEPTIRVTTFTMTCGYVWTCMVKSKDDVVSEESSNDEN 310
Query: 282 RLMTIVNT---RNRFNPPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQAT 338
L T R PP P Y+GNC T +L G+ G V + A +A
Sbjct: 311 ELEYFSFTADCRGLLTPPCPPNYFGNCLASCVAKATHKELVGDK-GLLVAVA--AIGEAI 367
Query: 339 EEYMHSMADFLVANRRCL-----FTTVRSCIVSDLTRFKLHETDFGWGEPVCGGVAKGGA 393
E+ +H+ L + L + R ++ +F + DFGWG+P AK
Sbjct: 368 EKRLHNEKGVLADAKTWLSESNGIPSKRFLGITGSPKFDSYGVDFGWGKP-----AKFDI 422
Query: 394 GLYGGASYIIACKNAKGEDGRVLVICLPVENMKRFAK 430
A I ++ E G + + LP +M FAK
Sbjct: 423 TSVDYAELIYVIQSRDFEKGVEIGVSLPKIHMDAFAK 459
>BACC_BACLI (O68008) Bacitracin synthetase 3 (BA3) [Includes:
ATP-dependent isoleucine adenylase (IleA) (Isoleucine
activase); ATP-dependent D-phenylalanine adenylase
(D-PheA) (D-phenylalanine activase); ATP-dependent
histidine adenylase (HisA) (Histidine
Length = 6359
Score = 38.9 bits (89), Expect = 0.026
Identities = 46/169 (27%), Positives = 71/169 (41%), Gaps = 22/169 (13%)
Query: 24 SSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQ-AIRQALSRTLVYYYPFA 82
SS + +L+ I+ GL +N+PF M + D D + A RQ + R F
Sbjct: 3591 SSAQRRLYILNQIEP-GGLSYNMPFAMKIAGD---FDVDRFEDAFRQLIERHEALRTAF- 3645
Query: 83 GRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVPGSEQVLD 142
+MVD GE V IE D V++G P EE + +
Sbjct: 3646 ----------VMVD--GEPVQKIEKEVDFK-VKYGRLGQDPL---EEKIKAFIKPFALEK 3689
Query: 143 SPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQP 191
+PLL +V + ++ L +H +SDG + F LAE+ +G + P
Sbjct: 3690 APLLRAEVLKASGDEHVLMLDMHHIISDGVSMAIFTRELAELYEGKTLP 3738
>GAS1_YEAST (P22146) Glycolipid anchored surface protein 1 precursor
(Glycoprotein GP115)
Length = 559
Score = 31.2 bits (69), Expect = 5.5
Identities = 19/43 (44%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Query: 362 SCIVSDLTRFKLHETDFGW--GEPVCGGV-AKGGAGLYGGASY 401
SC+VSD +ET F W E C G+ A G AG YG S+
Sbjct: 378 SCVVSDDVDSDDYETLFNWICNEVDCSGISANGTAGKYGAYSF 420
>YUXG_BACSU (P40747) Hypothetical oxidoreductase yuxG (EC 1.-.-.-)
(ORF2)
Length = 689
Score = 30.8 bits (68), Expect = 7.2
Identities = 17/44 (38%), Positives = 24/44 (53%), Gaps = 4/44 (9%)
Query: 391 GGAGLYGGASYIIACKNAKGEDGRVLVICLPVENMKRFAKELNN 434
GGAG G A AC+ E G V+V L +E ++ A E+N+
Sbjct: 434 GGAGGIGSA----ACRRFAAEGGHVIVADLNIEGAQKIAGEIND 473
>SRF2_BACSU (Q04747) Surfactin synthetase subunit 2
Length = 3587
Score = 30.8 bits (68), Expect = 7.2
Identities = 36/170 (21%), Positives = 68/170 (39%), Gaps = 21/170 (12%)
Query: 22 PASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTLVYYYPF 81
P SS + +L+ +D Q + +N+P +++ E D P + Q ++R
Sbjct: 2103 PLSSAQKRMYVLNQLDRQT-ISYNMPSVLLMEGE---LDIWPARLTPQLVNR-------- 2150
Query: 82 AGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVPGSEQVL 141
R ++ GE V I A+V L F +E + +
Sbjct: 2151 -----HESLRTSFMEANGEPVQRIIEKAEVDLHVFEAKEDEADQKIKEFIRPF----DLN 2201
Query: 142 DSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQP 191
D+PL+ + R++ ++ L +H ++DG F+ LA + +G P
Sbjct: 2202 DAPLIRAALLRIEAKKHLLLLDMHHIIADGVSRGIFVKELALLYKGEQLP 2251
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.140 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,052,348
Number of Sequences: 164201
Number of extensions: 2268178
Number of successful extensions: 4659
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 4604
Number of HSP's gapped (non-prelim): 18
length of query: 437
length of database: 59,974,054
effective HSP length: 113
effective length of query: 324
effective length of database: 41,419,341
effective search space: 13419866484
effective search space used: 13419866484
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0175.13


