

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
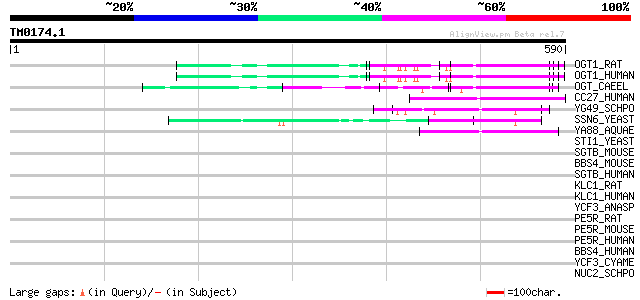
Query= TM0174.1
(590 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
OGT1_RAT (P56558) UDP-N-acetylglucosamine--peptide N-acetylgluco... 63 2e-09
OGT1_HUMAN (O15294) UDP-N-acetylglucosamine--peptide N-acetylglu... 63 2e-09
OGT_CAEEL (O18158) UDP-N-acetylglucosamine--peptide N-acetylgluc... 62 3e-09
CC27_HUMAN (P30260) Cell division cycle protein 27 homolog (CDC2... 50 2e-05
YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II 50 2e-05
SSN6_YEAST (P14922) Glucose repression mediator protein 50 2e-05
YA88_AQUAE (O67178) Hypothetical protein AQ_1088 46 2e-04
STI1_YEAST (P15705) Heat shock protein STI1 44 0.001
SGTB_MOUSE (Q8VD33) Small glutamine-rich tetratricopeptide repea... 44 0.001
BBS4_MOUSE (Q8C1Z7) Bardet-Biedl syndrome 4 protein homolog 44 0.001
SGTB_HUMAN (Q96EQ0) Small glutamine-rich tetratricopeptide repea... 43 0.002
KLC1_RAT (P37285) Kinesin light chain 1 (KLC 1) 43 0.002
KLC1_HUMAN (Q07866) Kinesin light chain 1 (KLC 1) 43 0.002
YCF3_ANASP (Q8YS98) Photosystem I assembly protein ycf3 42 0.003
PE5R_RAT (Q925N3) PEX5-related protein (Peroxin 5-related protei... 42 0.004
PE5R_MOUSE (Q8C437) PEX5-related protein (Peroxin 5-related prot... 42 0.004
PE5R_HUMAN (Q8IYB4) PEX5-related protein (Peroxin 5-related prot... 42 0.004
BBS4_HUMAN (Q96RK4) Bardet-Biedl syndrome 4 protein 42 0.006
YCF3_CYAME (Q85FT1) Photosystem I assembly protein ycf3 41 0.008
NUC2_SCHPO (P10505) Nuclear scaffold-like protein p76 41 0.008
>OGT1_RAT (P56558) UDP-N-acetylglucosamine--peptide
N-acetylglucosaminyltransferase 110 kDa subunit (EC
2.4.1.-) (O-GlcNAc transferase p110 subunit)
Length = 1036
Score = 62.8 bits (151), Expect = 2e-09
Identities = 100/418 (23%), Positives = 165/418 (38%), Gaps = 58/418 (13%)
Query: 178 AAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQE 237
A ++ LG + + G++Q AI L + P+ D NL A + G++E + + +
Sbjct: 79 AEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAY-- 136
Query: 238 LILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKAD 297
V+ + Y L C + + G L + AK C L AI+
Sbjct: 137 ---------VSALQYNPDLYCVRSDL----------GNLLKALGRLEEAKACYLKAIETQ 177
Query: 298 VKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSE 357
A W NL F+ G+ + EKA L+PN + Y + +KEA
Sbjct: 178 PNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDA-YINLGNVLKEAR------- 229
Query: 358 LLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEER--- 414
+A+ +R S LA V+ Q I A ++ + + E++
Sbjct: 230 ---IFDRAVAAYLR-ALSLSPNHAVVHGNLACVYYEQGLIDLAIDTYRRAI-ELQPHFPD 284
Query: 415 AVCSL------KQAVAEDPD---DPVRWHQLGVHSLCT-QQFKTSQKYLKAAVACDRGC- 463
A C+L K +VAE D +R SL K Q ++ AV R
Sbjct: 285 AYCNLANALKEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAVRLYRKAL 344
Query: 464 ------SYTWSNLGVSLQLSEEPSQAEKAYKQALLLA-TKQQAHAILSNLGIFYRHEKKY 516
+ SNL LQ + +A YK+A+ ++ T A+ SN+G + +
Sbjct: 345 EVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAY---SNMGNTLKEMQDV 401
Query: 517 QRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
Q A +T+++++ P +A A +NL + G + EA + AL+ P A NL
Sbjct: 402 QGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAIASYRTALKLKPDFPDAYCNL 459
Score = 57.4 bits (137), Expect = 1e-07
Identities = 39/115 (33%), Positives = 62/115 (53%), Gaps = 2/115 (1%)
Query: 469 NLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLE 528
NL +L + + A +AY AL + + S+LG + + + AKA + K++E
Sbjct: 118 NLAAALVAAGDMEGAVQAYVSALQY--NPDLYCVRSDLGNLLKALGRLEEAKACYLKAIE 175
Query: 529 LQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKI 583
QP +A A++NLG VF A+G + A + FEKA+ DP A NL V+ ++I
Sbjct: 176 TQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARI 230
Score = 53.1 bits (126), Expect = 2e-06
Identities = 58/234 (24%), Positives = 100/234 (41%), Gaps = 37/234 (15%)
Query: 383 AWAGLAMVHKAQHEI-----------------SSAYESEQHVLTEME--ERAVCSLKQAV 423
AW+ L V AQ EI AY + +VL E +RAV + +A+
Sbjct: 183 AWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVAAYLRAL 242
Query: 424 AEDPDDPVR-------WHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQL 476
+ P+ V +++ G+ L ++ A+ + NL +L+
Sbjct: 243 SLSPNHAVVHGNLACVYYEQGLIDLAIDTYRR-------AIELQPHFPDAYCNLANALKE 295
Query: 477 SEEPSQAEKAYKQALLLATKQQAHA-ILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAP 535
++AE Y AL L HA L+NL R + + A ++ K+LE+ P +A
Sbjct: 296 KGSVAEAEDCYNTALRLCP---THADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAA 352
Query: 536 AFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKICKGLLK 589
A +NL V +G L+EA +++A++ P A SN+ + + +G L+
Sbjct: 353 AHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQDVQGALQ 406
Score = 48.5 bits (114), Expect = 5e-05
Identities = 40/125 (32%), Positives = 55/125 (44%), Gaps = 8/125 (6%)
Query: 458 ACDRGCSYTW----SNLGVSLQLSEEPSQAEKAYKQALL--LATKQQA--HAILSNLGIF 509
A +R C W N GV L LS Q + + A LA KQ SNLG
Sbjct: 29 AAERHCMQLWRQEPDNTGVLLLLSSIHFQCRRLDRSAHFSTLAIKQNPLLAEAYSNLGNV 88
Query: 510 YRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDA 569
Y+ + Q A + +L L+P + + NL VA G +E A + ALQ +P L
Sbjct: 89 YKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAYVSALQYNPDLYC 148
Query: 570 AKSNL 574
+S+L
Sbjct: 149 VRSDL 153
Score = 47.8 bits (112), Expect = 8e-05
Identities = 49/218 (22%), Positives = 85/218 (38%), Gaps = 21/218 (9%)
Query: 380 LPTAWAGLAMVHKAQHEISSAYESEQHVLTEME-------------------ERAVCSLK 420
L A++ L V+K + ++ A E +H L E AV +
Sbjct: 78 LAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAYV 137
Query: 421 QAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEP 480
A+ +PD LG + + ++ A+ + WSNLG E
Sbjct: 138 SALQYNPDLYCVRSDLGNLLKALGRLEEAKACYLKAIETQPNFAVAWSNLGCVFNAQGEI 197
Query: 481 SQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNL 540
A +++A+ L I NLG + + + RA A + ++L L P +A NL
Sbjct: 198 WLAIHHFEKAVTLDPNFLDAYI--NLGNVLKEARIFDRAVAAYLRALSLSPNHAVVHGNL 255
Query: 541 GLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVV 578
V+ +GL++ A + +A++ P A NL +
Sbjct: 256 ACVYYEQGLIDLAIDTYRRAIELQPHFPDAYCNLANAL 293
Score = 42.7 bits (99), Expect = 0.003
Identities = 93/466 (19%), Positives = 170/466 (35%), Gaps = 85/466 (18%)
Query: 81 RKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIHHAQC 140
++ P A A+ LG +++ Q Q+AI Y A + +PD + I+ A
Sbjct: 73 KQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRL---------KPDFID-GYINLAAA 122
Query: 141 LILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISV 200
L+ E + + ++Q++ V + LG +L GR++ A +
Sbjct: 123 LVAAGDMEGA-------------VQAYVSALQYNPDLYCVRSDLGNLLKALGRLEEAKAC 169
Query: 201 LSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKY 260
+ P NLG + G + L+ F++ + D N A +N +L
Sbjct: 170 YLKAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVL---- 225
Query: 261 ASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSS 320
E + D+ +AA L A+ A + GNLA + G +
Sbjct: 226 -----------KEARIFDRAVAA------YLRALSLSPNHAVVHGNLACVYYEQGLIDLA 268
Query: 321 SKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVEL 380
+A +L+P+ Y + +KE + + +++L
Sbjct: 269 IDTYRRAIELQPH-FPDAYCNLANALKEKGSVAEAEDCY--------------NTALRLC 313
Query: 381 PT---AWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLG 437
PT + LA + + Q I A + L E A A
Sbjct: 314 PTHADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAAAHSNLA--------------- 358
Query: 438 VHSLCTQQFKTSQ--KYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLL-A 494
S+ QQ K + + K A+ + +SN+G +L+ ++ A + Y +A+ +
Sbjct: 359 --SVLQQQGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQDVQGALQCYTRAIQINP 416
Query: 495 TKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNL 540
AH SNL ++ A A + +L+L+P + A+ NL
Sbjct: 417 AFADAH---SNLASIHKDSGNIPEAIASYRTALKLKPDFPDAYCNL 459
>OGT1_HUMAN (O15294) UDP-N-acetylglucosamine--peptide
N-acetylglucosaminyltransferase 110 kDa subunit (EC
2.4.1.-) (O-GlcNAc transferase p110 subunit)
Length = 1036
Score = 62.8 bits (151), Expect = 2e-09
Identities = 100/418 (23%), Positives = 165/418 (38%), Gaps = 58/418 (13%)
Query: 178 AAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQE 237
A ++ LG + + G++Q AI L + P+ D NL A + G++E + + +
Sbjct: 79 AEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAY-- 136
Query: 238 LILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKAD 297
V+ + Y L C + + G L + AK C L AI+
Sbjct: 137 ---------VSALQYNPDLYCVRSDL----------GNLLKALGRLEEAKACYLKAIETQ 177
Query: 298 VKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSE 357
A W NL F+ G+ + EKA L+PN + Y + +KEA
Sbjct: 178 PNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDA-YINLGNVLKEAR------- 229
Query: 358 LLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEER--- 414
+A+ +R S LA V+ Q I A ++ + + E++
Sbjct: 230 ---IFDRAVAAYLR-ALSLSPNHAVVHGNLACVYYEQGLIDLAIDTYRRAI-ELQPHFPD 284
Query: 415 AVCSL------KQAVAEDPD---DPVRWHQLGVHSLCT-QQFKTSQKYLKAAVACDRGC- 463
A C+L K +VAE D +R SL K Q ++ AV R
Sbjct: 285 AYCNLANALKEKGSVAEAEDCYNTALRLCPTHADSLNNLANIKREQGNIEEAVRLYRKAL 344
Query: 464 ------SYTWSNLGVSLQLSEEPSQAEKAYKQALLLA-TKQQAHAILSNLGIFYRHEKKY 516
+ SNL LQ + +A YK+A+ ++ T A+ SN+G + +
Sbjct: 345 EVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAY---SNMGNTLKEMQDV 401
Query: 517 QRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
Q A +T+++++ P +A A +NL + G + EA + AL+ P A NL
Sbjct: 402 QGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAIASYRTALKLKPDFPDAYCNL 459
Score = 57.4 bits (137), Expect = 1e-07
Identities = 39/115 (33%), Positives = 62/115 (53%), Gaps = 2/115 (1%)
Query: 469 NLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLE 528
NL +L + + A +AY AL + + S+LG + + + AKA + K++E
Sbjct: 118 NLAAALVAAGDMEGAVQAYVSALQY--NPDLYCVRSDLGNLLKALGRLEEAKACYLKAIE 175
Query: 529 LQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKI 583
QP +A A++NLG VF A+G + A + FEKA+ DP A NL V+ ++I
Sbjct: 176 TQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARI 230
Score = 53.1 bits (126), Expect = 2e-06
Identities = 58/234 (24%), Positives = 100/234 (41%), Gaps = 37/234 (15%)
Query: 383 AWAGLAMVHKAQHEI-----------------SSAYESEQHVLTEME--ERAVCSLKQAV 423
AW+ L V AQ EI AY + +VL E +RAV + +A+
Sbjct: 183 AWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVAAYLRAL 242
Query: 424 AEDPDDPVR-------WHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQL 476
+ P+ V +++ G+ L ++ A+ + NL +L+
Sbjct: 243 SLSPNHAVVHGNLACVYYEQGLIDLAIDTYRR-------AIELQPHFPDAYCNLANALKE 295
Query: 477 SEEPSQAEKAYKQALLLATKQQAHA-ILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAP 535
++AE Y AL L HA L+NL R + + A ++ K+LE+ P +A
Sbjct: 296 KGSVAEAEDCYNTALRLCP---THADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAA 352
Query: 536 AFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKICKGLLK 589
A +NL V +G L+EA +++A++ P A SN+ + + +G L+
Sbjct: 353 AHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQDVQGALQ 406
Score = 48.5 bits (114), Expect = 5e-05
Identities = 40/125 (32%), Positives = 55/125 (44%), Gaps = 8/125 (6%)
Query: 458 ACDRGCSYTW----SNLGVSLQLSEEPSQAEKAYKQALL--LATKQQA--HAILSNLGIF 509
A +R C W N GV L LS Q + + A LA KQ SNLG
Sbjct: 29 AAERHCMQLWRQEPDNTGVLLLLSSIHFQCRRLDRSAHFSTLAIKQNPLLAEAYSNLGNV 88
Query: 510 YRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDA 569
Y+ + Q A + +L L+P + + NL VA G +E A + ALQ +P L
Sbjct: 89 YKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAYVSALQYNPDLYC 148
Query: 570 AKSNL 574
+S+L
Sbjct: 149 VRSDL 153
Score = 47.8 bits (112), Expect = 8e-05
Identities = 49/218 (22%), Positives = 85/218 (38%), Gaps = 21/218 (9%)
Query: 380 LPTAWAGLAMVHKAQHEISSAYESEQHVLTEME-------------------ERAVCSLK 420
L A++ L V+K + ++ A E +H L E AV +
Sbjct: 78 LAEAYSNLGNVYKERGQLQEAIEHYRHALRLKPDFIDGYINLAAALVAAGDMEGAVQAYV 137
Query: 421 QAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEP 480
A+ +PD LG + + ++ A+ + WSNLG E
Sbjct: 138 SALQYNPDLYCVRSDLGNLLKALGRLEEAKACYLKAIETQPNFAVAWSNLGCVFNAQGEI 197
Query: 481 SQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNL 540
A +++A+ L I NLG + + + RA A + ++L L P +A NL
Sbjct: 198 WLAIHHFEKAVTLDPNFLDAYI--NLGNVLKEARIFDRAVAAYLRALSLSPNHAVVHGNL 255
Query: 541 GLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVV 578
V+ +GL++ A + +A++ P A NL +
Sbjct: 256 ACVYYEQGLIDLAIDTYRRAIELQPHFPDAYCNLANAL 293
Score = 42.7 bits (99), Expect = 0.003
Identities = 93/466 (19%), Positives = 170/466 (35%), Gaps = 85/466 (18%)
Query: 81 RKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIHHAQC 140
++ P A A+ LG +++ Q Q+AI Y A + +PD + I+ A
Sbjct: 73 KQNPLLAEAYSNLGNVYKERGQLQEAIEHYRHALRL---------KPDFID-GYINLAAA 122
Query: 141 LILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISV 200
L+ E + + ++Q++ V + LG +L GR++ A +
Sbjct: 123 LVAAGDMEGA-------------VQAYVSALQYNPDLYCVRSDLGNLLKALGRLEEAKAC 169
Query: 201 LSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKY 260
+ P NLG + G + L+ F++ + D N A +N +L
Sbjct: 170 YLKAIETQPNFAVAWSNLGCVFNAQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVL---- 225
Query: 261 ASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSS 320
E + D+ +AA L A+ A + GNLA + G +
Sbjct: 226 -----------KEARIFDRAVAA------YLRALSLSPNHAVVHGNLACVYYEQGLIDLA 268
Query: 321 SKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVEL 380
+A +L+P+ Y + +KE + + +++L
Sbjct: 269 IDTYRRAIELQPH-FPDAYCNLANALKEKGSVAEAEDCY--------------NTALRLC 313
Query: 381 PT---AWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLG 437
PT + LA + + Q I A + L E A A
Sbjct: 314 PTHADSLNNLANIKREQGNIEEAVRLYRKALEVFPEFAAAHSNLA--------------- 358
Query: 438 VHSLCTQQFKTSQ--KYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLL-A 494
S+ QQ K + + K A+ + +SN+G +L+ ++ A + Y +A+ +
Sbjct: 359 --SVLQQQGKLQEALMHYKEAIRISPTFADAYSNMGNTLKEMQDVQGALQCYTRAIQINP 416
Query: 495 TKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNL 540
AH SNL ++ A A + +L+L+P + A+ NL
Sbjct: 417 AFADAH---SNLASIHKDSGNIPEAIASYRTALKLKPDFPDAYCNL 459
>OGT_CAEEL (O18158) UDP-N-acetylglucosamine--peptide
N-acetylglucosaminyltransferase (EC 2.4.1.-) (O-GlcNAc)
(OGT)
Length = 1151
Score = 62.4 bits (150), Expect = 3e-09
Identities = 67/288 (23%), Positives = 119/288 (41%), Gaps = 31/288 (10%)
Query: 291 LAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAE 350
+ AIK + + A + NL + G + + + + A KL+P
Sbjct: 183 MLAIKVNNQCAEAYSNLGNYYKEKGQLQDALENYKLAVKLKP------------------ 224
Query: 351 RSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTE 410
E + N A+++ GD L + TA+ ++ + + S + +
Sbjct: 225 ------EFIDAYINLAAALVSGGD--LEQAVTAYFNALQINPDLYCVRSDLGNLLKAMGR 276
Query: 411 MEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNL 470
+EE VC LK A+ P V W LG + + + + AV D + NL
Sbjct: 277 LEEAKVCYLK-AIETQPQFAVAWSNLGCVFNSQGEIWLAIHHFEKAVTLDPNFLDAYINL 335
Query: 471 GVSLQLSEEPSQAEKAYKQALLLATKQQAHAIL-SNLGIFYRHEKKYQRAKAMFTKSLEL 529
G L+ + +A AY +AL L+ HA++ NL Y + A + K+++L
Sbjct: 336 GNVLKEARIFDRAVSAYLRALNLSGN---HAVVHGNLACVYYEQGLIDLAIDTYKKAIDL 392
Query: 530 QPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKV 577
QP + A+ NL +G + EA+ + KAL+ P +++NL +
Sbjct: 393 QPHFPDAYCNLANALKEKGSVVEAEQMYMKALELCPTHADSQNNLANI 440
Score = 61.2 bits (147), Expect = 7e-09
Identities = 94/439 (21%), Positives = 168/439 (37%), Gaps = 61/439 (13%)
Query: 142 ILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVL 201
+L S+ N K LE +L+ ++ + + A ++ LG + G++Q A+
Sbjct: 162 LLLLSAINFQTKNLEKSMQYSMLA-----IKVNNQCAEAYSNLGNYYKEKGQLQDALENY 216
Query: 202 SSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYA 261
+ + PE D NL A + G+LE + + + + + L C +
Sbjct: 217 KLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAYFNALQINPD-----------LYCVRS 265
Query: 262 SVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSS 321
+ G L + AK C L AI+ + A W NL F+ G+ +
Sbjct: 266 DL----------GNLLKAMGRLEEAKVCYLKAIETQPQFAVAWSNLGCVFNSQGEIWLAI 315
Query: 322 KCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELP 381
EKA L+PN + Y + +KEA + ++ G+ ++V
Sbjct: 316 HHFEKAVTLDPNFLDA-YINLGNVLKEARIFDRAVSAY------LRALNLSGNHAVVH-- 366
Query: 382 TAWAGLAMVHKAQHEISSAYESEQHVLTEMEE--RAVCSLKQAVAEDPDDPVRWHQLGVH 439
LA V+ Q I A ++ + + A C+L A+ E
Sbjct: 367 ---GNLACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGS----------- 412
Query: 440 SLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATK-QQ 498
+ Q Y+KA C + +NL + + A + Y +AL + +
Sbjct: 413 -----VVEAEQMYMKALELCPTHAD-SQNNLANIKREQGKIEDATRLYLKALEIYPEFAA 466
Query: 499 AHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFE 558
AH SNL + + K A + +++ + P +A A++N+G G A C+
Sbjct: 467 AH---SNLASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMGNTLKEMGDSSAAIACYN 523
Query: 559 KALQSDPLLDAAKSNLVKV 577
+A+Q +P A SNL +
Sbjct: 524 RAIQINPAFADAHSNLASI 542
Score = 55.8 bits (133), Expect = 3e-07
Identities = 38/115 (33%), Positives = 61/115 (53%), Gaps = 2/115 (1%)
Query: 469 NLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLE 528
NL +L + QA AY AL + + + S+LG + + + AK + K++E
Sbjct: 232 NLAAALVSGGDLEQAVTAYFNALQI--NPDLYCVRSDLGNLLKAMGRLEEAKVCYLKAIE 289
Query: 529 LQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKI 583
QP +A A++NLG VF ++G + A + FEKA+ DP A NL V+ ++I
Sbjct: 290 TQPQFAVAWSNLGCVFNSQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARI 344
Score = 52.8 bits (125), Expect = 3e-06
Identities = 51/190 (26%), Positives = 90/190 (46%), Gaps = 14/190 (7%)
Query: 394 QHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLG-----VHSLCTQQFKT 448
Q +++ A +Q +L ++ SL Q +A P V + + V L +QF+
Sbjct: 83 QFQLNGATAVQQQLLLTPQQ----SLAQPIALAPQPTVVLNGVSETLKKVTELAHRQFQ- 137
Query: 449 SQKYLKAAVACDRGCSYTWSNLGVSLQLSE---EPSQAEKAYKQALL-LATKQQAHAILS 504
S Y++A C+ +NL L LS + EK+ + ++L + Q S
Sbjct: 138 SGNYVEAEKYCNLVFQSDPNNLPTLLLLSAINFQTKNLEKSMQYSMLAIKVNNQCAEAYS 197
Query: 505 NLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSD 564
NLG +Y+ + + Q A + +++L+P + A+ NL V+ G LE+A + ALQ +
Sbjct: 198 NLGNYYKEKGQLQDALENYKLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAYFNALQIN 257
Query: 565 PLLDAAKSNL 574
P L +S+L
Sbjct: 258 PDLYCVRSDL 267
Score = 45.4 bits (106), Expect = 4e-04
Identities = 32/109 (29%), Positives = 56/109 (51%), Gaps = 4/109 (3%)
Query: 467 WSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHA-ILSNLGIFYRHEKKYQRAKAMFTK 525
+ NL +L+ +AE+ Y +AL L HA +NL R + K + A ++ K
Sbjct: 400 YCNLANALKEKGSVVEAEQMYMKALELCP---THADSQNNLANIKREQGKIEDATRLYLK 456
Query: 526 SLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
+LE+ P +A A +NL + +G L +A +++A++ P A SN+
Sbjct: 457 ALEIYPEFAAAHSNLASILQQQGKLNDAILHYKEAIRIAPTFADAYSNM 505
Score = 43.5 bits (101), Expect = 0.002
Identities = 62/279 (22%), Positives = 109/279 (38%), Gaps = 9/279 (3%)
Query: 307 LAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVAT--HRMKEAERSQDPSELLSFGGN 364
LA+ SG++ + K + +PN + T ++ + K E+S S L N
Sbjct: 131 LAHRQFQSGNYVEAEKYCNLVFQSDPNNLPTLLLLSAINFQTKNLEKSMQYSMLAIKVNN 190
Query: 365 EMASIIRDGDSSLVE---LPTAWAGLAMVHKAQHEISSAYESEQHVLTEME--ERAVCSL 419
+ A + + E L A + K + E AY + L E+AV +
Sbjct: 191 QCAEAYSNLGNYYKEKGQLQDALENYKLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAY 250
Query: 420 KQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEE 479
A+ +PD LG + + ++ A+ + WSNLG E
Sbjct: 251 FNALQINPDLYCVRSDLGNLLKAMGRLEEAKVCYLKAIETQPQFAVAWSNLGCVFNSQGE 310
Query: 480 PSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNN 539
A +++A+ L I NLG + + + RA + + ++L L +A N
Sbjct: 311 IWLAIHHFEKAVTLDPNFLDAYI--NLGNVLKEARIFDRAVSAYLRALNLSGNHAVVHGN 368
Query: 540 LGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVV 578
L V+ +GL++ A ++KA+ P A NL +
Sbjct: 369 LACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANAL 407
Score = 32.7 bits (73), Expect = 2.7
Identities = 39/205 (19%), Positives = 80/205 (39%), Gaps = 21/205 (10%)
Query: 140 CLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAIS 199
C S N ++ + E ++++ ++++ AA + L IL + G++ AI
Sbjct: 427 CPTHADSQNNLANIKREQGKIEDATRLYLKALEIYPEFAAAHSNLASILQQQGKLNDAIL 486
Query: 200 VLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCK 259
+ IAP D N+G ++G+ + C+ I Q +P ++ L
Sbjct: 487 HYKEAIRIAPTFADAYSNMGNTLKEMGDSSAAIACYNRAI---QINPAFADAHSNL---- 539
Query: 260 YASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRS 319
A G +A+ + + + A + +K D A+ NLA+ I D
Sbjct: 540 -------ASIHKDAGNMAEAIQSYSTALK-----LKPDFPDAYC--NLAHCHQIICDWND 585
Query: 320 SSKCLEKAAKLEPNCMSTRYAVATH 344
K + K ++ + + + + H
Sbjct: 586 YDKRVRKLVQIVEDQLCKKRLPSVH 610
>CC27_HUMAN (P30260) Cell division cycle protein 27 homolog
(CDC27Hs) (H-NUC)
Length = 824
Score = 50.1 bits (118), Expect = 2e-05
Identities = 38/165 (23%), Positives = 75/165 (45%), Gaps = 2/165 (1%)
Query: 426 DPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEK 485
D + P W G ++ + K+ + A+ D +Y ++ LG L+EE +A
Sbjct: 563 DKNSPEAWCAAGNCFSLQREHDIAIKFFQRAIQVDPNYAYAYTLLGHEFVLTEELDKALA 622
Query: 486 AYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFV 545
++ A+ + + + LG+ Y ++K+ A+ F K+L++ P + ++G+V
Sbjct: 623 CFRNAIRVNPRH--YNAWYGLGMIYYKQEKFSLAEMHFQKALDINPQSSVLLCHIGVVQH 680
Query: 546 AEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKICKGLLKE 590
A E+A KA+ DP K + V+ ++ K L+E
Sbjct: 681 ALKKSEKALDTLNKAIVIDPKNPLCKFHRASVLFANEKYKSALQE 725
Score = 34.7 bits (78), Expect = 0.72
Identities = 31/161 (19%), Positives = 69/161 (42%), Gaps = 2/161 (1%)
Query: 405 QHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCS 464
+ VLTE ++A+ + A+ +P W+ LG+ ++F ++ + + A+ + S
Sbjct: 610 EFVLTEELDKALACFRNAIRVNPRHYNAWYGLGMIYYKQEKFSLAEMHFQKALDINPQSS 669
Query: 465 YTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFT 524
++GV ++ +A +A+++ K + + +E KY+ A
Sbjct: 670 VLLCHIGVVQHALKKSEKALDTLNKAIVIDPKNPL-CKFHRASVLFANE-KYKSALQELE 727
Query: 525 KSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDP 565
+ ++ P + + +G V+ G A F A+ DP
Sbjct: 728 ELKQIVPKESLVYFLIGKVYKKLGQTHLALMNFSWAMDLDP 768
Score = 32.3 bits (72), Expect = 3.6
Identities = 56/290 (19%), Positives = 109/290 (37%), Gaps = 54/290 (18%)
Query: 185 GFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQN 244
G++ L + + AI++LS L + L +G AY ++ + + F E + + +N
Sbjct: 472 GYLALCSYNCKEAINILSHLPSHHYNTGWVLCQIGRAYFELSEYMQAERIFSE-VRRIEN 530
Query: 245 HPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIW 304
+ V + + L VA S L D D S W
Sbjct: 531 YRVEGMEIYSTTLWHLQKDVA---LSVLSKDLTDM-----------------DKNSPEAW 570
Query: 305 GNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGN 364
FS+ +H + K ++A +++PN + Y + H E +
Sbjct: 571 CAAGNCFSLQREHDIAIKFFQRAIQVDPN-YAYAYTLLGHEFVLTEEL-----------D 618
Query: 365 EMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVL---------------- 408
+ + R+ AW GL M++ Q + S A Q L
Sbjct: 619 KALACFRNAIRVNPRHYNAWYGLGMIYYKQEKFSLAEMHFQKALDINPQSSVLLCHIGVV 678
Query: 409 ---TEMEERAVCSLKQAVAEDPDDPV-RWHQLGVHSLCTQQFKTSQKYLK 454
+ E+A+ +L +A+ DP +P+ ++H+ V +++K++ + L+
Sbjct: 679 QHALKKSEKALDTLNKAIVIDPKNPLCKFHRASV-LFANEKYKSALQELE 727
>YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II
Length = 1102
Score = 49.7 bits (117), Expect = 2e-05
Identities = 33/161 (20%), Positives = 71/161 (43%), Gaps = 3/161 (1%)
Query: 408 LTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTW 467
L + +++A+ + + A+ ++P Q+ +QF + +Y + + CD W
Sbjct: 346 LFDDQDKALSAYESALRQNPYSIPAMLQIATILRNREQFPLAIEYYQTILDCDPKQGEIW 405
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSL 527
S LG + ++ S+A AY+QAL + + +GI Y ++ A+ F + L
Sbjct: 406 SALGHCYLMQDDLSRAYSAYRQALYHLKDPKDPKLWYGIGILYDRYGSHEHAEEAFMQCL 465
Query: 528 ELQPGYAPA---FNNLGLVFVAEGLLEEAKYCFEKALQSDP 565
+ P + + LG+++ + ++ F L + P
Sbjct: 466 RMDPNFEKVNEIYFRLGIIYKQQHKFAQSLELFRHILDNPP 506
Score = 46.6 bits (109), Expect = 2e-04
Identities = 47/217 (21%), Positives = 97/217 (44%), Gaps = 33/217 (15%)
Query: 387 LAMVHKAQHEISSAYESEQHVLTE-----------------MEERAVCSL-----KQAVA 424
L +++K QH+ + + E +H+L E+R L ++ +A
Sbjct: 481 LGIIYKQQHKFAQSLELFRHILDNPPKPLTVLDIYFQIGHVYEQRKEYKLAKEAYERVLA 540
Query: 425 EDPDDPVRWHQLGVHSLCTQQFK--TSQ----KYLKAAVACDRGCSYTWSNLGVSLQLSE 478
E P+ QLG LC QQ T+Q +YL ++ D + +W +G +
Sbjct: 541 ETPNHAKVLQQLGW--LCHQQSSSFTNQDLAIQYLTKSLEADDTDAQSWYLIGRCYVAQQ 598
Query: 479 EPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFN 538
+ ++A +AY+QA+ + ++G+ Y +YQ A +++++ L P + +
Sbjct: 599 KYNKAYEAYQQAVYRDGRNPT--FWCSIGVLYYQINQYQDALDAYSRAIRLNPYISEVWY 656
Query: 539 NLGLVFVA-EGLLEEAKYCFEKALQSDPLLDAAKSNL 574
+LG ++ + + +A +++A + DP K+ L
Sbjct: 657 DLGTLYESCHNQISDALDAYQRAAELDPTNPHIKARL 693
>SSN6_YEAST (P14922) Glucose repression mediator protein
Length = 966
Score = 49.7 bits (117), Expect = 2e-05
Identities = 75/348 (21%), Positives = 130/348 (36%), Gaps = 49/348 (14%)
Query: 170 SVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLE 229
++QF+ A +L + Q A + L + PE D LG YL + +L+
Sbjct: 72 TLQFNPSSAKALTSLAHLYRSRDMFQRAAELYERALLVNPELSDVWATLGHCYLMLDDLQ 131
Query: 230 LSAKCFQELI--LKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANV-- 285
+ +Q+ + L + N P L + +L +Y S+ A A L AN
Sbjct: 132 RAYNAYQQALYHLSNPNVP-KLWHGIGILYDRYGSLDYAEEAFAKVLELDPHFEKANEIY 190
Query: 286 ---------------AKEC---LLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKA 327
A EC +L A ++ IW L G+ + + + E
Sbjct: 191 FRLGIIYKHQGKWSQALECFRYILPQPPAPLQEWDIWFQLGSVLESMGEWQGAKEAYEHV 250
Query: 328 -AKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAG 386
A+ + + + + M + DP + L + + S+ D + T W
Sbjct: 251 LAQNQHHAKVLQQLGCLYGMSNVQ-FYDPQKALDY---LLKSLEADPSDA-----TTWYH 301
Query: 387 LAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQF 446
L VH + + ++AY++ Q QAV D +P+ W +GV Q+
Sbjct: 302 LGRVHMIRTDYTAAYDAFQ---------------QAVNRDSRNPIFWCSIGVLYYQISQY 346
Query: 447 KTSQKYLKAAVACDRGCSYTWSNLGVSLQ-LSEEPSQAEKAYKQALLL 493
+ + A+ + S W +LG + + + S A AYKQA L
Sbjct: 347 RDALDAYTRAIRLNPYISEVWYDLGTLYETCNNQLSDALDAYKQAARL 394
Score = 47.8 bits (112), Expect = 8e-05
Identities = 31/123 (25%), Positives = 54/123 (43%), Gaps = 3/123 (2%)
Query: 446 FKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSN 505
F+ + + + A+ + S W+ LG + ++ +A AY+QAL + +
Sbjct: 96 FQRAAELYERALLVNPELSDVWATLGHCYLMLDDLQRAYNAYQQALYHLSNPNVPKLWHG 155
Query: 506 LGIFYRHEKKYQRAKAMFTKSLELQPGYAPA---FNNLGLVFVAEGLLEEAKYCFEKALQ 562
+GI Y A+ F K LEL P + A + LG+++ +G +A CF L
Sbjct: 156 IGILYDRYGSLDYAEEAFAKVLELDPHFEKANEIYFRLGIIYKHQGKWSQALECFRYILP 215
Query: 563 SDP 565
P
Sbjct: 216 QPP 218
Score = 33.1 bits (74), Expect = 2.1
Identities = 30/124 (24%), Positives = 56/124 (44%), Gaps = 8/124 (6%)
Query: 444 QQFKTSQKYLKAAVA------CDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQ 497
QQ + Q+ +AAV + + TW ++ + + +A AY A L
Sbjct: 20 QQQQQQQQQQQAAVPQQPLDPLTQSTAETWLSIASLAETLGDGDRAAMAY-DATLQFNPS 78
Query: 498 QAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCF 557
A A L++L YR +QRA ++ ++L + P + + LG ++ L+ A +
Sbjct: 79 SAKA-LTSLAHLYRSRDMFQRAAELYERALLVNPELSDVWATLGHCYLMLDDLQRAYNAY 137
Query: 558 EKAL 561
++AL
Sbjct: 138 QQAL 141
>YA88_AQUAE (O67178) Hypothetical protein AQ_1088
Length = 761
Score = 46.2 bits (108), Expect = 2e-04
Identities = 34/149 (22%), Positives = 72/149 (47%), Gaps = 3/149 (2%)
Query: 436 LGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLAT 495
LG++ + + + +++ L + S+LG+ AE+ K+AL +
Sbjct: 41 LGIYHFLSGEPQKAEELLSQVSENSLNSAQGLSDLGLLYFFLGRVEDAERVLKKALKFSD 100
Query: 496 KQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKY 555
A + + LG Y + K + A+ + ++L L P NLG++ + +G LE+A
Sbjct: 101 VDDA--LYARLGALYYSQGKLEEAQHYWERALSLNPNKVEILYNLGVLHLNKGELEKALD 158
Query: 556 CFEKALQSDP-LLDAAKSNLVKVVTMSKI 583
FE+AL+ P +A + + ++++++I
Sbjct: 159 LFERALRLKPDFREAEEKKTLILLSLNRI 187
Score = 36.6 bits (83), Expect = 0.19
Identities = 31/160 (19%), Positives = 72/160 (44%), Gaps = 2/160 (1%)
Query: 406 HVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSY 465
H L+ ++A L Q + LG+ + + +++ LK A+
Sbjct: 45 HFLSGEPQKAEELLSQVSENSLNSAQGLSDLGLLYFFLGRVEDAERVLKKALKFSDVDDA 104
Query: 466 TWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTK 525
++ LG + +A+ +++AL L + IL NLG+ + ++ + ++A +F +
Sbjct: 105 LYARLGALYYSQGKLEEAQHYWERALSLNPNKVE--ILYNLGVLHLNKGELEKALDLFER 162
Query: 526 SLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDP 565
+L L+P + A L+ ++ ++E + + L+ +P
Sbjct: 163 ALRLKPDFREAEEKKTLILLSLNRIDELVEEYYRELEKNP 202
>STI1_YEAST (P15705) Heat shock protein STI1
Length = 589
Score = 43.9 bits (102), Expect = 0.001
Identities = 30/99 (30%), Positives = 48/99 (48%), Gaps = 6/99 (6%)
Query: 485 KAYKQALLLATKQ------QAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFN 538
K Y +A+ L TK H + SN Y KK+ A + +++ P ++ +N
Sbjct: 19 KDYDKAIELFTKAIEVSETPNHVLYSNRSACYTSLKKFSDALNDANECVKINPSWSKGYN 78
Query: 539 NLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKV 577
LG + G L+EA+ ++KAL+ D AAK L +V
Sbjct: 79 RLGAAHLGLGDLDEAESNYKKALELDASNKAAKEGLDQV 117
>SGTB_MOUSE (Q8VD33) Small glutamine-rich tetratricopeptide
repeat-containing protein B
Length = 304
Score = 43.9 bits (102), Expect = 0.001
Identities = 31/113 (27%), Positives = 52/113 (45%), Gaps = 7/113 (6%)
Query: 469 NLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFT---- 524
++G + QL +E + K A + QA + N ++Y + Q + +T
Sbjct: 81 DVGKADQLKDEGNNHMKEENYAAAVDCYTQAIELDPNNAVYYCNRAAAQSKLSHYTDAIK 140
Query: 525 ---KSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
K++ + Y+ A+ +GL A EEA ++KAL DP D+ KSNL
Sbjct: 141 DCEKAIAIDSKYSKAYGRMGLALTAMNKFEEAVTSYQKALDLDPENDSYKSNL 193
Score = 33.5 bits (75), Expect = 1.6
Identities = 34/150 (22%), Positives = 62/150 (40%), Gaps = 7/150 (4%)
Query: 402 ESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDR 461
E H+ E AV QA+ DP++ V + + + K + A+A D
Sbjct: 91 EGNNHMKEENYAAAVDCYTQAIELDPNNAVYYCNRAAAQSKLSHYTDAIKDCEKAIAIDS 150
Query: 462 GCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKA 521
S + +G++L + +A +Y++AL L + ++ SNL I E+K + +
Sbjct: 151 KYSKAYGRMGLALTAMNKFEEAVTSYQKALDLDPENDSYK--SNLKI---AEQKLREVSS 205
Query: 522 MFTKSLELQPGYAPAFNNLGLVFVAEGLLE 551
L A NN + +A L++
Sbjct: 206 PTGTGLSFD--MASLINNPAFITMAASLMQ 233
>BBS4_MOUSE (Q8C1Z7) Bardet-Biedl syndrome 4 protein homolog
Length = 520
Score = 43.9 bits (102), Expect = 0.001
Identities = 48/231 (20%), Positives = 91/231 (38%), Gaps = 57/231 (24%)
Query: 77 IHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIH 136
+H A ++ K+ + +LG +H KAI +Y+KA E E +LL+ + +
Sbjct: 158 LHSALQLNKHDLTYIMLGKIHLLQGDLDKAIEIYKKAVEF------SPENTELLTTLGLL 211
Query: 137 HAQCLILESSSENSSDK-ELEPHELKEIL---SKLKESVQFDI-------------RQAA 179
+ Q + + + E+ + +P K IL S ++ FD+
Sbjct: 212 YLQLGVYQKAFEHLGNALTYDPANYKAILAAGSMMQTHGDFDVALTKYRVVACAIPESPP 271
Query: 180 VWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYL---------------- 223
+WN +G + +AIS L +AP ++ L NLG+ +L
Sbjct: 272 LWNNIGMCFFGKKKYVAAISCLKRANYLAPFDWKILYNLGLVHLTMQQYASAFHFLSAAI 331
Query: 224 ------------------QIGNLELSAKCFQELILKDQNHPVALVNYAALL 256
+ ++E + + + E + D+ +P+ +NYA LL
Sbjct: 332 NFQPKMGELYMLLAVALTNLEDIENARRAYVEAVRLDKCNPLVNLNYAVLL 382
Score = 39.7 bits (91), Expect = 0.022
Identities = 38/149 (25%), Positives = 59/149 (39%), Gaps = 21/149 (14%)
Query: 184 LGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQ 243
LG I L G + AI + + +PEN + L LG+ YLQ+G + K F+ L
Sbjct: 174 LGKIHLLQGDLDKAIEIYKKAVEFSPENTELLTTLGLLYLQLG---VYQKAFEHLGNALT 230
Query: 244 NHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHI 303
P NY A+L AG+ D + C + +S +
Sbjct: 231 YDP---ANYKAIL---------AAGSMMQTHGDFDVALTKYRVVACAIP------ESPPL 272
Query: 304 WGNLAYAFSISGDHRSSSKCLEKAAKLEP 332
W N+ F + ++ CL++A L P
Sbjct: 273 WNNIGMCFFGKKKYVAAISCLKRANYLAP 301
Score = 37.7 bits (86), Expect = 0.085
Identities = 60/285 (21%), Positives = 112/285 (39%), Gaps = 25/285 (8%)
Query: 308 AYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMA 367
A F + G+ + S + + A L P C VA + + + +E+ NE A
Sbjct: 74 ALIFRLEGNIQESLELFQTCAVLSPQCADNLKQVA-RSLFLLGKHKAATEVY----NEAA 128
Query: 368 SIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEME-----------ERAV 416
+ + L + L +KAQ ++ SA + +H LT + ++A+
Sbjct: 129 KLNQKDWEICHNLGVCYTYLKQFNKAQDQLHSALQLNKHDLTYIMLGKIHLLQGDLDKAI 188
Query: 417 CSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQL 476
K+AV P++ LG+ L ++ + ++L A+ D G +Q
Sbjct: 189 EIYKKAVEFSPENTELLTTLGLLYLQLGVYQKAFEHLGNALTYDPANYKAILAAGSMMQT 248
Query: 477 SEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPA 536
+ A Y+ ++ ++ + +N+G+ + +KKY A + ++ L P
Sbjct: 249 HGDFDVALTKYR--VVACAIPESPPLWNNIGMCFFGKKKYVAAISCLKRANYLAPFDWKI 306
Query: 537 FNNLGLVFVAEGLLEEAKYCFEKALQSDP-------LLDAAKSNL 574
NLGLV + A + A+ P LL A +NL
Sbjct: 307 LYNLGLVHLTMQQYASAFHFLSAAINFQPKMGELYMLLAVALTNL 351
>SGTB_HUMAN (Q96EQ0) Small glutamine-rich tetratricopeptide
repeat-containing protein B (Small glutamine-rich
protein with tetratricopeptide repeats 2)
Length = 304
Score = 43.1 bits (100), Expect = 0.002
Identities = 31/113 (27%), Positives = 51/113 (44%), Gaps = 7/113 (6%)
Query: 469 NLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFT---- 524
++G + QL +E + K A + QA + N ++Y + Q +T
Sbjct: 81 DVGKADQLKDEGNNHMKEENYAAAVDCYTQAIELDPNNAVYYCNRAAAQSKLGHYTDAIK 140
Query: 525 ---KSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
K++ + Y+ A+ +GL A EEA ++KAL DP D+ KSNL
Sbjct: 141 DCEKAIAIDSKYSKAYGRMGLALTALNKFEEAVTSYQKALDLDPENDSYKSNL 193
>KLC1_RAT (P37285) Kinesin light chain 1 (KLC 1)
Length = 556
Score = 43.1 bits (100), Expect = 0.002
Identities = 27/90 (30%), Positives = 45/90 (50%), Gaps = 16/90 (17%)
Query: 501 AILSNLGIFYRHEKKYQRAKAMFTKSLELQ--------PGYAPAFNNLGLVFVAEGLLEE 552
A L+NL + Y KY+ A+ + ++LE++ P A NNL L+ +G EE
Sbjct: 294 ATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAKQLNNLALLCQNQGKYEE 353
Query: 553 AKYCFEKALQ--------SDPLLDAAKSNL 574
+Y +++AL+ DP + K+NL
Sbjct: 354 VEYYYQRALEIYQTKLGPDDPNVAKTKNNL 383
Score = 33.1 bits (74), Expect = 2.1
Identities = 34/134 (25%), Positives = 55/134 (40%), Gaps = 22/134 (16%)
Query: 450 QKYLKAAVACDRGCSY-------TWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAH-- 500
Q++ AA A +G Y T NL + A KQAL K H
Sbjct: 189 QQHSSAAAAAQQG-GYEIPARLRTLHNLVIQYASQGRYEVAVPLCKQALEDLEKTSGHDH 247
Query: 501 ----AILSNLGIFYRHEKKYQRAKAMFTKSLELQ--------PGYAPAFNNLGLVFVAEG 548
+L+ L + YR + KY+ A + +L ++ P A NNL +++ G
Sbjct: 248 PDVATMLNILALVYRDQNKYKDAANLLNDALAIREKTLGRDHPAVAATLNNLAVLYGKRG 307
Query: 549 LLEEAKYCFEKALQ 562
+EA+ ++AL+
Sbjct: 308 KYKEAEPLCKRALE 321
>KLC1_HUMAN (Q07866) Kinesin light chain 1 (KLC 1)
Length = 569
Score = 43.1 bits (100), Expect = 0.002
Identities = 27/90 (30%), Positives = 45/90 (50%), Gaps = 16/90 (17%)
Query: 501 AILSNLGIFYRHEKKYQRAKAMFTKSLELQ--------PGYAPAFNNLGLVFVAEGLLEE 552
A L+NL + Y KY+ A+ + ++LE++ P A NNL L+ +G EE
Sbjct: 294 ATLNNLAVLYGKRGKYKEAEPLCKRALEIREKVLGKDHPDVAKQLNNLALLCQNQGKYEE 353
Query: 553 AKYCFEKALQ--------SDPLLDAAKSNL 574
+Y +++AL+ DP + K+NL
Sbjct: 354 VEYYYQRALEIYQTKLGPDDPNVAKTKNNL 383
Score = 33.1 bits (74), Expect = 2.1
Identities = 34/134 (25%), Positives = 55/134 (40%), Gaps = 22/134 (16%)
Query: 450 QKYLKAAVACDRGCSY-------TWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAH-- 500
Q++ AA A +G Y T NL + A KQAL K H
Sbjct: 189 QQHSSAAAAAQQG-GYEIPARLRTLHNLVIQYASQGRYEVAVPLCKQALEDLEKTSGHDH 247
Query: 501 ----AILSNLGIFYRHEKKYQRAKAMFTKSLELQ--------PGYAPAFNNLGLVFVAEG 548
+L+ L + YR + KY+ A + +L ++ P A NNL +++ G
Sbjct: 248 PDVATMLNILALVYRDQNKYKDAANLLNDALAIREKTLGKDHPAVAATLNNLAVLYGKRG 307
Query: 549 LLEEAKYCFEKALQ 562
+EA+ ++AL+
Sbjct: 308 KYKEAEPLCKRALE 321
>YCF3_ANASP (Q8YS98) Photosystem I assembly protein ycf3
Length = 173
Score = 42.4 bits (98), Expect = 0.003
Identities = 29/112 (25%), Positives = 56/112 (49%), Gaps = 4/112 (3%)
Query: 471 GVSLQLSEEPSQAEKAYKQALLLATKQQAHA-ILSNLGIFYRHEKKYQRAKAMFTKSLEL 529
G+S Q E ++A + Y++AL L IL N+G+ Y + +A ++ +++EL
Sbjct: 42 GMSAQAEGEYAEALEYYEEALTLEEDTNDRGYILYNMGLIYASNGDHDKALELYHQAIEL 101
Query: 530 QPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMS 581
P A NN+ +++ +G E+AK + + L D A ++ + M+
Sbjct: 102 NPRLPQALNNIAVIYHYQG--EKAKETGDHD-GGEALFDQAADYWIRAIRMA 150
>PE5R_RAT (Q925N3) PEX5-related protein (Peroxin 5-related protein)
(Pex5Rp) (TPR-containing Rab8b-interacting protein)
Length = 602
Score = 42.0 bits (97), Expect = 0.004
Identities = 28/94 (29%), Positives = 50/94 (52%), Gaps = 2/94 (2%)
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSL 527
+ LGV LS E ++A A+ AL + + + +++ + LG + + + A +T++L
Sbjct: 454 TGLGVLFHLSGEFNRAIDAFNAALTV--RPEDYSLWNRLGATLANGDRSEEAVEAYTRAL 511
Query: 528 ELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKAL 561
E+QPG+ + NLG+ + G EA F AL
Sbjct: 512 EIQPGFIRSRYNLGISCINLGAYREAVSNFLTAL 545
Score = 31.2 bits (69), Expect = 7.9
Identities = 20/99 (20%), Positives = 41/99 (41%)
Query: 153 KELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENY 212
K L+ +L + ++ ++ D A W LG + Q+AI L L + P N
Sbjct: 311 KRLKEGDLPVTILFMEAAILQDPGNAEAWQFLGITQAENENEQAAIVALQRCLELQPNNL 370
Query: 213 DCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVN 251
L L ++Y + + + + + I ++ + + N
Sbjct: 371 KALMALAVSYTNTSHQQDACEALKNWIKQNPKYKYLVKN 409
>PE5R_MOUSE (Q8C437) PEX5-related protein (Peroxin 5-related
protein) (Pex5Rp) (PEX2-related protein)
Length = 567
Score = 42.0 bits (97), Expect = 0.004
Identities = 28/94 (29%), Positives = 50/94 (52%), Gaps = 2/94 (2%)
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSL 527
+ LGV LS E ++A A+ AL + + + +++ + LG + + + A +T++L
Sbjct: 419 TGLGVLFHLSGEFNRAIDAFNAALTV--RPEDYSLWNRLGATLANGDRSEEAVEAYTRAL 476
Query: 528 ELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKAL 561
E+QPG+ + NLG+ + G EA F AL
Sbjct: 477 EIQPGFIRSRYNLGISCINLGAYREAVSNFLTAL 510
Score = 31.2 bits (69), Expect = 7.9
Identities = 20/99 (20%), Positives = 41/99 (41%)
Query: 153 KELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENY 212
K L+ +L + ++ ++ D A W LG + Q+AI L L + P N
Sbjct: 276 KRLKEGDLPVTILFMEAAILQDPGDAEAWQFLGITQAENENEQAAIVALQRCLELQPNNL 335
Query: 213 DCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVN 251
L L ++Y + + + + + I ++ + + N
Sbjct: 336 KALMALAVSYTNTSHQQDACEALKNWIKQNPKYKYLVKN 374
>PE5R_HUMAN (Q8IYB4) PEX5-related protein (Peroxin 5-related
protein) (Pex5Rp) (PEX2-related protein)
Length = 626
Score = 42.0 bits (97), Expect = 0.004
Identities = 28/94 (29%), Positives = 50/94 (52%), Gaps = 2/94 (2%)
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSL 527
+ LGV LS E ++A A+ AL + + + +++ + LG + + + A +T++L
Sbjct: 478 TGLGVLFHLSGEFNRAIDAFNAALTV--RPEDYSLWNRLGATLANGDRSEEAVEAYTRAL 535
Query: 528 ELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKAL 561
E+QPG+ + NLG+ + G EA F AL
Sbjct: 536 EIQPGFIRSRYNLGISCINLGAYREAVSNFLTAL 569
Score = 32.3 bits (72), Expect = 3.6
Identities = 38/182 (20%), Positives = 66/182 (35%), Gaps = 13/182 (7%)
Query: 153 KELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENY 212
K L+ +L + ++ ++ D A W LG + Q+AI L L + P N
Sbjct: 335 KRLKEGDLPVTILFMEAAILQDPGDAEAWQFLGITQAENENEQAAIVALQRCLELQPNNL 394
Query: 213 DCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASAS 272
L L ++Y G+ + + + I ++ Y L+ K S G S
Sbjct: 395 KALMALAVSYTNTGHQQDACDALKNWIKQNP-------KYKYLVKSKKGS--PGLTRRMS 445
Query: 273 EGALADQVMAANVAKECLLAAI--KADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKL 330
+ + V+ KE L A D+ + L F +SG+ + A +
Sbjct: 446 KSPVDSSVLEG--VKELYLEAAHQNGDMIDPDLQTGLGVLFHLSGEFNRAIDAFNAALTV 503
Query: 331 EP 332
P
Sbjct: 504 RP 505
>BBS4_HUMAN (Q96RK4) Bardet-Biedl syndrome 4 protein
Length = 519
Score = 41.6 bits (96), Expect = 0.006
Identities = 40/164 (24%), Positives = 70/164 (42%), Gaps = 23/164 (14%)
Query: 77 IHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIH 136
+H A + ++ + +LG +H KAI VY+KA E E +LL+ + +
Sbjct: 158 LHNALNLNRHDLTYIMLGKIHLLEGDLDKAIEVYKKAVEF------SPENTELLTTLGLL 211
Query: 137 HAQCLILESSSENSSDK-ELEPHELKEIL---SKLKESVQFDI-------------RQAA 179
+ Q I + + E+ + +P K IL S ++ FD+
Sbjct: 212 YLQLGIYQKAFEHLGNALTYDPTNYKAILAAGSMMQTHGDFDVALTKYRVVACAVPESPP 271
Query: 180 VWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYL 223
+WN +G + +AIS L +AP ++ L NLG+ +L
Sbjct: 272 LWNNIGMCFFGKKKYVAAISCLKRANYLAPFDWKILYNLGLVHL 315
Score = 40.0 bits (92), Expect = 0.017
Identities = 40/149 (26%), Positives = 59/149 (38%), Gaps = 21/149 (14%)
Query: 184 LGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQ 243
LG I L G + AI V + +PEN + L LG+ YLQ+G + K F+ L
Sbjct: 174 LGKIHLLEGDLDKAIEVYKKAVEFSPENTELLTTLGLLYLQLG---IYQKAFEHLGNALT 230
Query: 244 NHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHI 303
P NY A+L AG+ D + C A +S +
Sbjct: 231 YDP---TNYKAIL---------AAGSMMQTHGDFDVALTKYRVVAC------AVPESPPL 272
Query: 304 WGNLAYAFSISGDHRSSSKCLEKAAKLEP 332
W N+ F + ++ CL++A L P
Sbjct: 273 WNNIGMCFFGKKKYVAAISCLKRANYLAP 301
Score = 32.0 bits (71), Expect = 4.7
Identities = 23/79 (29%), Positives = 39/79 (49%)
Query: 415 AVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSL 474
A+ LK+A P D + LG+ L QQ+ ++ +L AA+ + L V+L
Sbjct: 289 AISCLKRANYLAPFDWKILYNLGLVHLTMQQYASAFHFLSAAINFQPKMGELYMLLAVAL 348
Query: 475 QLSEEPSQAEKAYKQALLL 493
E+ A++AY +A+ L
Sbjct: 349 TNLEDTENAKRAYAEAVHL 367
>YCF3_CYAME (Q85FT1) Photosystem I assembly protein ycf3
Length = 158
Score = 41.2 bits (95), Expect = 0.008
Identities = 28/110 (25%), Positives = 56/110 (50%), Gaps = 8/110 (7%)
Query: 464 SYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHA-ILSNLGIFYRHEKKYQRAKAM 522
++++ G++ Q E +QA ++Y AL + I+ N+G+ Y + ++A
Sbjct: 35 AFSYYREGMAAQAEGEYAQALESYYHALEFEEDVIDRSYIIYNIGLIYASNGEDEQALEY 94
Query: 523 FTKSLELQPGYAPAFNNLGLVFVAEG-------LLEEAKYCFEKALQSDP 565
+ ++LEL P A NN+ +++ +G LL++A + KA+Q P
Sbjct: 95 YHQALELNPRLVQALNNIAVIYHKQGMTYQDEQLLQKAAAYWRKAIQLAP 144
>NUC2_SCHPO (P10505) Nuclear scaffold-like protein p76
Length = 665
Score = 41.2 bits (95), Expect = 0.008
Identities = 26/101 (25%), Positives = 53/101 (51%), Gaps = 4/101 (3%)
Query: 466 TWSNLGVSLQLSEEPSQAEKAYKQALLL-ATKQQAHAILSNLGIFYRHEKKYQRAKAMFT 524
+W L L E SQA K +A+ L T + A+ + G + ++Y+++K F
Sbjct: 433 SWCILANCFSLQREHSQALKCINRAIQLDPTFEYAYTLQ---GHEHSANEEYEKSKTSFR 489
Query: 525 KSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDP 565
K++ + + A+ LG+V++ G ++A + F++A + +P
Sbjct: 490 KAIRVNVRHYNAWYGLGMVYLKTGRNDQADFHFQRAAEINP 530
Score = 35.0 bits (79), Expect = 0.55
Identities = 42/186 (22%), Positives = 71/186 (37%), Gaps = 25/186 (13%)
Query: 219 GIAYLQIGNLELSAKCFQELILKDQNHPVAL----VNYAALLLCKYASVVAGAGASASEG 274
G+ L L + CFQ L ++ QN P L + Y L+ + + V S
Sbjct: 336 GVYLLAQYKLREALNCFQSLPIEQQNTPFVLAKLGITYFELVDYEKSEEVFQKLRDLSPS 395
Query: 275 ALADQVMAA----NVAKECLLA-----AIKADVKSAHIWGNLAYAFSISGDHRSSSKCLE 325
+ D + + ++ K L+ ++ + S W LA FS+ +H + KC+
Sbjct: 396 RVKDMEVFSTALWHLQKSVPLSYLAHETLETNPYSPESWCILANCFSLQREHSQALKCIN 455
Query: 326 KAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWA 385
+A +L+P + YA + + + SF R V AW
Sbjct: 456 RAIQLDP---TFEYAYTLQGHEHSANEEYEKSKTSF---------RKAIRVNVRHYNAWY 503
Query: 386 GLAMVH 391
GL MV+
Sbjct: 504 GLGMVY 509
Score = 34.7 bits (78), Expect = 0.72
Identities = 44/209 (21%), Positives = 80/209 (38%), Gaps = 18/209 (8%)
Query: 53 DVDQHVQGAPSSREEKVSSMKTGLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEK 112
+V Q ++ SR + + T L H+ + +P + AH L P Y
Sbjct: 384 EVFQKLRDLSPSRVKDMEVFSTALWHLQKSVPLSYLAHETLE------TNP------YSP 431
Query: 113 AEEILLRPETEIERPDLLSLVQIHHAQCLILESSSENSSDKELEPHELKEILSKLKES-- 170
+L ++R +L I+ A + L+ + E + + H E K K S
Sbjct: 432 ESWCILANCFSLQREHSQALKCINRA--IQLDPTFEYAYTLQGHEHSANEEYEKSKTSFR 489
Query: 171 --VQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNL 228
++ ++R W LG + LKTGR A I P N + +G+ Y + +
Sbjct: 490 KAIRVNVRHYNAWYGLGMVYLKTGRNDQADFHFQRAAEINPNNSVLITCIGMIYERCKDY 549
Query: 229 ELSAKCFQELILKDQNHPVALVNYAALLL 257
+ + + D+ +A A +L+
Sbjct: 550 KKALDFYDRACKLDEKSSLARFKKAKVLI 578
Score = 32.7 bits (73), Expect = 2.7
Identities = 36/163 (22%), Positives = 69/163 (42%), Gaps = 24/163 (14%)
Query: 232 AKCFQELILKDQNHPVALVNYAALL--LCKYASVVAGAGASASEGALADQVMAANVAKEC 289
A CF + ++ + + +N A L +YA + G SA+E +K
Sbjct: 438 ANCFS--LQREHSQALKCINRAIQLDPTFEYAYTLQGHEHSANE--------EYEKSKTS 487
Query: 290 LLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPN--CMSTRYAVATHRMK 347
AI+ +V+ + W L + +G + + ++AA++ PN + T + R K
Sbjct: 488 FRKAIRVNVRHYNAWYGLGMVYLKTGRNDQADFHFQRAAEINPNNSVLITCIGMIYERCK 547
Query: 348 EAERSQD----------PSELLSFGGNEMASIIRDGDSSLVEL 380
+ +++ D S L F ++ ++ D D +LVEL
Sbjct: 548 DYKKALDFYDRACKLDEKSSLARFKKAKVLILLHDHDKALVEL 590
Score = 32.3 bits (72), Expect = 3.6
Identities = 26/160 (16%), Positives = 68/160 (42%), Gaps = 2/160 (1%)
Query: 421 QAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEP 480
+ + +P P W L ++ + K + A+ D Y ++ G +EE
Sbjct: 422 ETLETNPYSPESWCILANCFSLQREHSQALKCINRAIQLDPTFEYAYTLQGHEHSANEEY 481
Query: 481 SQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNL 540
+++ ++++A+ + + + LG+ Y + +A F ++ E+ P + +
Sbjct: 482 EKSKTSFRKAIRVNVRH--YNAWYGLGMVYLKTGRNDQADFHFQRAAEINPNNSVLITCI 539
Query: 541 GLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTM 580
G+++ ++A +++A + D A+ KV+ +
Sbjct: 540 GMIYERCKDYKKALDFYDRACKLDEKSSLARFKKAKVLIL 579
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.129 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 63,946,733
Number of Sequences: 164201
Number of extensions: 2520744
Number of successful extensions: 8117
Number of sequences better than 10.0: 141
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 83
Number of HSP's that attempted gapping in prelim test: 7605
Number of HSP's gapped (non-prelim): 509
length of query: 590
length of database: 59,974,054
effective HSP length: 116
effective length of query: 474
effective length of database: 40,926,738
effective search space: 19399273812
effective search space used: 19399273812
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0174.1


