

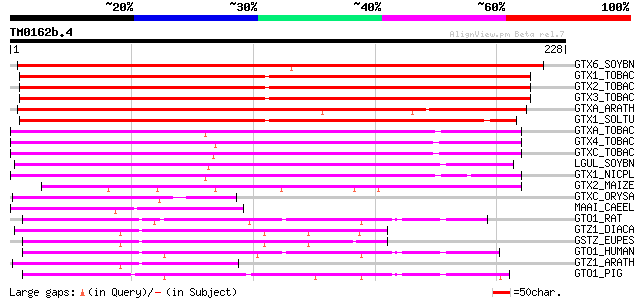
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0162b.4
(228 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
GTX6_SOYBN (P32110) Probable glutathione S-transferase (EC 2.5.1... 223 2e-58
GTX1_TOBAC (Q03662) Probable glutathione S-transferase (EC 2.5.1... 216 5e-56
GTX2_TOBAC (Q03663) Probable glutathione S-transferase (EC 2.5.1... 209 3e-54
GTX3_TOBAC (Q03664) Probable glutathione S-transferase (EC 2.5.1... 208 7e-54
GTXA_ARATH (P46421) Glutathione S-transferase 103-1A (EC 2.5.1.18) 199 5e-51
GTX1_SOLTU (P32111) Probable glutathione S-transferase (EC 2.5.1... 180 2e-45
GTXA_TOBAC (P25317) Probable glutathione S-transferase parA (EC ... 169 5e-42
GTX4_TOBAC (Q03666) Probable glutathione S-transferase (EC 2.5.1... 169 5e-42
GTXC_TOBAC (P49332) Probable glutathione S-transferase parC (EC ... 167 1e-41
LGUL_SOYBN (P46417) Lactoylglutathione lyase (EC 4.4.1.5) (Methy... 160 2e-39
GTX1_NICPL (P50471) Probable glutathione S-transferase MSR-1 (EC... 158 1e-38
GTX2_MAIZE (P50472) Probable glutathione S-transferase BZ2 (EC 2... 113 4e-25
GTXC_ORYSA (Q06398) Probable glutathione S-transferase (EC 2.5.1... 77 4e-14
MAAI_CAEEL (Q18938) Probable maleylacetoacetate isomerase (EC 5.... 75 2e-13
GTO1_RAT (Q9Z339) Glutathione transferase omega 1 (EC 2.5.1.18) ... 74 2e-13
GTZ1_DIACA (P28342) Glutathione S-transferase 1 (EC 2.5.1.18) (S... 74 4e-13
GSTZ_EUPES (P57108) Glutathione S-transferase zeta class (EC 2.5... 71 2e-12
GTO1_HUMAN (P78417) Glutathione transferase omega 1 (EC 2.5.1.18... 70 5e-12
GTZ1_ARATH (Q9ZVQ3) Glutathione S-transferase zeta-class 1 (EC 2... 69 1e-11
GTO1_PIG (Q9N1F5) Glutathione transferase omega 1 (EC 2.5.1.18) ... 65 1e-10
>GTX6_SOYBN (P32110) Probable glutathione S-transferase (EC
2.5.1.18) (Heat shock protein 26A) (G2-4)
Length = 225
Score = 223 bits (569), Expect = 2e-58
Identities = 106/217 (48%), Positives = 149/217 (67%), Gaps = 1/217 (0%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
+DVKLL SPF RV+ ALKLKGVEY+++EE++ NKS LLL+ NPVHKK+PV VH +
Sbjct: 6 EDVKLLGIVGSPFVCRVQIALKLKGVEYKFLEENLGNKSDLLLKYNPVHKKVPVFVHNEQ 65
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAM-CITGDEQE 122
IAES +I+EYIDETWK P+LP DPY+RALARFW+ F + K++ V ++ + E+E
Sbjct: 66 PIAESLVIVEYIDETWKNNPILPSDPYQRALARFWSKFIDDKIVGAVSKSVFTVDEKERE 125
Query: 123 KALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFP 182
K + E EA++ +E ++ K+FFGGE G +DIA +I++WI I++E+ + + KFP
Sbjct: 126 KNVEETYEALQFLENELKDKKFFGGEEFGLVDIAAVFIAFWIPIFQEIAGLQLFTSEKFP 185
Query: 183 ATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLS 219
W FL+HP + + LPPRD + YF R + LS
Sbjct: 186 ILYKWSQEFLNHPFVHEVLPPRDPLFAYFKARYESLS 222
>GTX1_TOBAC (Q03662) Probable glutathione S-transferase (EC
2.5.1.18) (Auxin-induced protein PGNT1/PCNT110)
Length = 223
Score = 216 bits (549), Expect = 5e-56
Identities = 99/210 (47%), Positives = 145/210 (68%), Gaps = 1/210 (0%)
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
+VKLL FW SPFS+RVEWALK+KGV+YEY+EED NKSSLLL+ NP+HKK+PVL+H K
Sbjct: 3 EVKLLGFWYSPFSRRVEWALKIKGVKYEYIEEDRDNKSSLLLQSNPIHKKVPVLIHNGKR 62
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
I ES +I+EYIDET++ +LP DPY+RALARFWA F + K + V G+EQEK
Sbjct: 63 IVESMVILEYIDETFEGPSILPKDPYDRALARFWAKFLDDK-VPAVVKTFLRKGEEQEKD 121
Query: 125 LNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPAT 184
E E ++ ++ ++ K+FF G+ G+ DIA +++W+ ++EE + ++ KFP
Sbjct: 122 KEEVCEMLKVLDNELKDKKFFVGDKFGFADIAANLVAFWLGVFEEASGVVLVTSEKFPNF 181
Query: 185 TAWMTNFLSHPVIKDTLPPRDKMIDYFHGR 214
W +++ IK++LPPRD+++ ++ R
Sbjct: 182 CKWRGEYINCSQIKESLPPRDELLAFYRSR 211
>GTX2_TOBAC (Q03663) Probable glutathione S-transferase (EC
2.5.1.18) (Auxin-induced protein PGNT35/PCNT111)
Length = 223
Score = 209 bits (533), Expect = 3e-54
Identities = 99/210 (47%), Positives = 142/210 (67%), Gaps = 1/210 (0%)
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
+VKLL FW SPFS RVEWALK+KGV+YEY+EED NKSSLLL+ NPV+KK+PVL+H K
Sbjct: 3 EVKLLGFWYSPFSHRVEWALKIKGVKYEYIEEDRDNKSSLLLQSNPVYKKVPVLIHNGKP 62
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
I ES II+EYIDET++ +LP DPY+RALARFWA F + K + V G+EQEK
Sbjct: 63 IVESMIILEYIDETFEGPSILPKDPYDRALARFWAKFLDDK-VAAVVNTFFRKGEEQEKG 121
Query: 125 LNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPAT 184
E E ++ ++ ++ K+FF G+ G+ DIA + +W+ ++EE ++ KFP
Sbjct: 122 KEEVYEMLKVLDNELKDKKFFAGDKFGFADIAANLVGFWLGVFEEGYGDVLVKSEKFPNF 181
Query: 185 TAWMTNFLSHPVIKDTLPPRDKMIDYFHGR 214
+ W +++ + ++LPPRD+++ +F R
Sbjct: 182 SKWRDEYINCSQVNESLPPRDELLAFFRAR 211
>GTX3_TOBAC (Q03664) Probable glutathione S-transferase (EC
2.5.1.18) (Auxin-induced protein PCNT103)
Length = 223
Score = 208 bits (530), Expect = 7e-54
Identities = 98/210 (46%), Positives = 142/210 (66%), Gaps = 1/210 (0%)
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
+VKLL FW SPF+ RVEWALKLKGV+YEY+EED NKSSLLL+ NPVHKK+PVL+H K
Sbjct: 3 EVKLLGFWYSPFTHRVEWALKLKGVKYEYIEEDRDNKSSLLLQSNPVHKKVPVLIHNGKP 62
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
I ES +I+EYIDET++ +LP DPY+RALARFW+ F K + V G+EQEK
Sbjct: 63 IVESMVILEYIDETFEGPSILPKDPYDRALARFWSKFLGDK-VAAVVNTFFRKGEEQEKG 121
Query: 125 LNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPAT 184
E E ++ ++ ++ K+FF G+ G+ DIA + +W+ ++EE + ++ KFP
Sbjct: 122 KEEVYEMLKVLDNELKDKKFFVGDKFGFADIAANLVGFWLGVFEEGYGVVLVTSEKFPNF 181
Query: 185 TAWMTNFLSHPVIKDTLPPRDKMIDYFHGR 214
+ W +++ +K++LP RD+++ +F R
Sbjct: 182 SRWRDEYINCSQVKESLPSRDELLAFFRAR 211
>GTXA_ARATH (P46421) Glutathione S-transferase 103-1A (EC 2.5.1.18)
Length = 224
Score = 199 bits (506), Expect = 5e-51
Identities = 97/212 (45%), Positives = 142/212 (66%), Gaps = 4/212 (1%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
++VKLL W SPFS+RVE ALKLKG+ YEYVEE + NKS LLL LNP+HKK+PVLVH K
Sbjct: 5 EEVKLLGIWASPFSRRVEMALKLKGIPYEYVEEILENKSPLLLALNPIHKKVPVLVHNGK 64
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
TI ES +I+EYIDETW Q P+LP DPYER+ ARF+A +++++ F++M ++ +
Sbjct: 65 TILESHVILEYIDETWPQNPILPQDPYERSKARFFAKLVDEQIMNVGFISMARADEKGRE 124
Query: 124 ALNE-AREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWI--HIWEEVGSIHIIDPLK 180
L E RE + +E+ + GK +FGG+ +G+LD G + + WE +G + +I K
Sbjct: 125 VLAEQVRELIMYLEKELVGKDYFGGKTVGFLDFVAGSLIPFCLERGWEGIG-LEVITEEK 183
Query: 181 FPATTAWMTNFLSHPVIKDTLPPRDKMIDYFH 212
FP W+ N ++KD +PPR++ +++ +
Sbjct: 184 FPEFKRWVRNLEKVEIVKDCVPPREEHVEHMN 215
>GTX1_SOLTU (P32111) Probable glutathione S-transferase (EC
2.5.1.18) (Pathogenesis-related protein 1)
Length = 217
Score = 180 bits (457), Expect = 2e-45
Identities = 93/204 (45%), Positives = 131/204 (63%), Gaps = 3/204 (1%)
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
+VKLL SPFS RVEWALK+KGV+YE++EED+ NKS LLL+ NP+HKKIPVL+H K
Sbjct: 3 EVKLLGLRYSPFSHRVEWALKIKGVKYEFIEEDLQNKSPLLLQSNPIHKKIPVLIHNGKC 62
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
I ES +I+EYIDE ++ +LP DPY+RALARFWA + E K V+ + G+EQEKA
Sbjct: 63 ICESMVILEYIDEAFEGPSILPKDPYDRALARFWAKYVEDK-GAAVWKSFFSKGEEQEKA 121
Query: 125 LNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPAT 184
EA E ++ ++ + K+ F G+ G+ DI + ++ I EEV I + KFP
Sbjct: 122 KEEAYEMLKILDNEFKDKKCFVGDKFGFADIVANGAALYLGILEEVSGIVLATSEKFPNF 181
Query: 185 TAWMTNFLSHPVIKDTLPPRDKMI 208
AW + + ++ P RD+++
Sbjct: 182 CAWRDEYCTQN--EEYFPSRDELL 203
>GTXA_TOBAC (P25317) Probable glutathione S-transferase parA (EC
2.5.1.18) (Auxin-regulated protein parA) (STR246C
protein)
Length = 220
Score = 169 bits (428), Expect = 5e-42
Identities = 86/211 (40%), Positives = 128/211 (59%), Gaps = 3/211 (1%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
M S +V LL FWPS F R+ AL LKG++YE EE++ +KS LLLE+NPVHKKIP+L+H
Sbjct: 1 MESNNVVLLDFWPSSFGMRLRIALALKGIKYEAKEENLSDKSPLLLEMNPVHKKIPILIH 60
Query: 61 GHKTIAESFIIIEYIDETW-KQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGD 119
K I ES I+EYIDE W + PLLP DPYER+ ARFWA++ ++K+ T G+
Sbjct: 61 NSKAICESLNILEYIDEVWHDKCPLLPSDPYERSQARFWADYIDKKIYSTGRRVWSGKGE 120
Query: 120 EQEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPL 179
+QE+A E E ++ +E + K +FGG+N+G++D+A+ + W + +E + I
Sbjct: 121 DQEEAKKEFIEILKTLEGELGNKTYFGGDNLGFVDVALVPFTSWFYSYETCANFSI--EA 178
Query: 180 KFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
+ P W + + +LP K+ +
Sbjct: 179 ECPKLVVWAKTCMESESVSKSLPHPHKIYGF 209
>GTX4_TOBAC (Q03666) Probable glutathione S-transferase (EC
2.5.1.18) (Auxin-induced protein PCNT107)
Length = 221
Score = 169 bits (428), Expect = 5e-42
Identities = 86/212 (40%), Positives = 128/212 (59%), Gaps = 4/212 (1%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
M +++V LL FWPS F R+ AL K ++YEY EED+ NKS LLL++NP+HKKIPVL+H
Sbjct: 1 MANEEVILLDFWPSMFGMRLRIALAEKEIKYEYKEEDLRNKSPLLLQMNPIHKKIPVLIH 60
Query: 61 GHKTIAESFIIIEYIDETWKQYP--LLPHDPYERALARFWANFTEQKLLETVFVAMCITG 118
K I ES I +EYI+E WK LLP DPY+RA ARFWA++ ++KL + G
Sbjct: 61 NGKPICESIIAVEYIEEVWKDKAPNLLPSDPYDRAQARFWADYIDKKLYDFGRKLWTTKG 120
Query: 119 DEQEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDP 178
+EQE A + E ++ +E + K +FGGE+ G++DIA+ W + +E G+
Sbjct: 121 EEQEAAKKDFIECLKVLEGALGDKPYFGGESFGFVDIALIGYYSWFYAYETFGNFS--TE 178
Query: 179 LKFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
+ P AW + + +LP + K++++
Sbjct: 179 AECPKFVAWAKRCMQRESVAKSLPDQPKVLEF 210
>GTXC_TOBAC (P49332) Probable glutathione S-transferase parC (EC
2.5.1.18) (Auxin-regulated protein parC)
Length = 221
Score = 167 bits (424), Expect = 1e-41
Identities = 84/212 (39%), Positives = 128/212 (59%), Gaps = 4/212 (1%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
M +++V LL FWPS F R+ AL K ++YEY +ED+ NKS LLL++NP+HKKIPVL+H
Sbjct: 1 MANEEVILLDFWPSMFGMRLRIALAEKEIKYEYKQEDLRNKSPLLLQMNPIHKKIPVLIH 60
Query: 61 GHKTIAESFIIIEYIDETWKQY--PLLPHDPYERALARFWANFTEQKLLETVFVAMCITG 118
K I ES I +EYI+E WK LLP DPY+RA ARFWA++ ++KL + G
Sbjct: 61 NGKPICESIIAVEYIEEVWKDKAPSLLPSDPYDRAQARFWADYIDKKLYDFGRKLWATKG 120
Query: 119 DEQEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDP 178
+EQE A + E ++ +E + + +FGGE+ G++DIA+ W + +E G+
Sbjct: 121 EEQEAAKKDFIECLKVLEGALGDRPYFGGESFGFVDIALIGFYSWFYAYETFGNFS--TE 178
Query: 179 LKFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
+ P AW + + +LP + K++++
Sbjct: 179 AECPKFVAWAKRCMQRESVAKSLPDQPKVLEF 210
>LGUL_SOYBN (P46417) Lactoylglutathione lyase (EC 4.4.1.5)
(Methylglyoxalase) (Aldoketomutase) (Glyoxalase I)
Length = 219
Score = 160 bits (406), Expect = 2e-39
Identities = 78/206 (37%), Positives = 122/206 (58%), Gaps = 3/206 (1%)
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
S +V LL W S + R AL KGV YEY EE++ N+S LLL++NP+HKKIPVL+H
Sbjct: 2 SDEVVLLDTWASMYGMRARIALAEKGVRYEYKEENLMNRSPLLLQMNPIHKKIPVLIHNG 61
Query: 63 KTIAESFIIIEYIDETWK-QYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQ 121
K I ES II++YIDE W + PL+P DPY+R+ ARFW ++ ++K+ +T G+E
Sbjct: 62 KPICESAIIVQYIDEVWNDKSPLMPSDPYKRSQARFWVDYIDKKIYDTWKKMWLSKGEEH 121
Query: 122 EKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
E+ E +++EE + K F+G + G++D+ + S W + +E G+ + + +
Sbjct: 122 EEGKKELISIFKQLEETLTDKPFYGDDTFGFVDLCLITFSSWFYTYETYGNFKMEE--EC 179
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKM 207
P AW+ + + +TLP K+
Sbjct: 180 PKLMAWVKRCMERETVSNTLPDAKKV 205
>GTX1_NICPL (P50471) Probable glutathione S-transferase MSR-1 (EC
2.5.1.18) (Auxin-regulated protein MSR-1)
Length = 219
Score = 158 bits (399), Expect = 1e-38
Identities = 85/211 (40%), Positives = 127/211 (59%), Gaps = 4/211 (1%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
M S +V LL F S F R+ AL LKG++YE EE++ +KS LLLE+NPVHKKIP+L+H
Sbjct: 1 MESNNVVLLDFSGSSFGMRLRIALALKGIKYEAKEENLSDKSPLLLEMNPVHKKIPILIH 60
Query: 61 GHKTIAESFIIIEYIDETW-KQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGD 119
K I ES I+EYIDE W ++ PLLP DPY+R+ ARFWAN+ + K+ T G+
Sbjct: 61 NGKPICESLNILEYIDEVWHEKCPLLPSDPYQRSQARFWANYIDNKIYSTGRRVWSGKGE 120
Query: 120 EQEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPL 179
+QE+A E E + +E + K +FGG+N+G++D+A+ + W + +E + I
Sbjct: 121 DQEEAKKEFIEIFKTLEGELGNKTYFGGDNLGFVDVALVPFTSWFYSYETCANFSI--EA 178
Query: 180 KFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
+ W N + + + +LP K+ D+
Sbjct: 179 ECRKLVVWQ-NCMENERVSKSLPHPHKIYDF 208
>GTX2_MAIZE (P50472) Probable glutathione S-transferase BZ2 (EC
2.5.1.18) (Bronze-2 protein)
Length = 236
Score = 113 bits (282), Expect = 4e-25
Identities = 72/213 (33%), Positives = 111/213 (51%), Gaps = 16/213 (7%)
Query: 14 SPFSKRVEWALKLKGVEYEYVEEDIF-NKSSLLLELNPVHKKIPVLV-HGHKTIAESFII 71
SPF+ R AL L+GV YE ++E + KS LL NPV+ KIPVL+ + I ES +I
Sbjct: 9 SPFTARARLALDLRGVAYELLDEPLGPKKSDRLLAANPVYGKIPVLLLPDGRAICESAVI 68
Query: 72 IEYIDETWKQYP-------LLPHDPYERALARFWANFTEQKLLETV-FVAMCITGDEQEK 123
++YI++ ++ LLP DPYERA+ RFW F + K + V++ T + +
Sbjct: 69 VQYIEDVARESGGAEAGSLLLPDDPYERAMHRFWTAFIDDKFWPALDAVSLAPTPGARAQ 128
Query: 124 ALNEAREAMEKIEEVIE----GKRFFGGENI--GYLDIAVGWISYWIHIWEEVGSIHIID 177
A + R A+ +EE + G+ FF G + G LD+A+G + E + + +ID
Sbjct: 129 AAEDTRAALSLLEEAFKDRSNGRAFFSGGDAAPGLLDLALGCFLPALRACERLHGLSLID 188
Query: 178 PLKFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
P W F +HP K LP +K++ +
Sbjct: 189 ASATPLLDGWSQRFAAHPAAKRVLPDTEKVVQF 221
>GTXC_ORYSA (Q06398) Probable glutathione S-transferase (EC
2.5.1.18) (28 kDa cold-induced protein)
Length = 254
Score = 76.6 bits (187), Expect = 4e-14
Identities = 42/100 (42%), Positives = 58/100 (58%), Gaps = 13/100 (13%)
Query: 2 GSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH- 60
GS ++KLL W SP++ RV L LK + YEYVEE++ +KS LLL NPVHK +PVL+H
Sbjct: 3 GSGELKLLGVWSSPYAIRVRVVLNLKSLPYEYVEENLGDKSDLLLASNPVHKSVPVLLHA 62
Query: 61 -------GHKTIAESFIIIEYIDETWKQYPLLPHDPYERA 93
GH+ + + + ++P DPYERA
Sbjct: 63 GRRERVAGHRAVHR-----RGLAGARRGRSVMPSDPYERA 97
>MAAI_CAEEL (Q18938) Probable maleylacetoacetate isomerase (EC
5.2.1.2) (MAAI)
Length = 214
Score = 74.7 bits (182), Expect = 2e-13
Identities = 45/98 (45%), Positives = 57/98 (57%), Gaps = 3/98 (3%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKS--SLLLELNPVHKKIPVL 58
M +Q L S+W S S RV AL LK V+YEY D+ ++ S L E+NP K+P
Sbjct: 1 MSNQKPVLYSYWRSSCSWRVRIALALKNVDYEYKTVDLLSEEAKSKLKEINPA-AKVPTF 59
Query: 59 VHGHKTIAESFIIIEYIDETWKQYPLLPHDPYERALAR 96
V + I ES IIEY++ET PLLP DP +RA AR
Sbjct: 60 VVDGQVITESLAIIEYLEETHPDVPLLPKDPIKRAHAR 97
>GTO1_RAT (Q9Z339) Glutathione transferase omega 1 (EC 2.5.1.18)
(GSTO 1-1) (Glutathione-dependent dehydroascorbate
reductase)
Length = 241
Score = 74.3 bits (181), Expect = 2e-13
Identities = 59/196 (30%), Positives = 90/196 (45%), Gaps = 13/196 (6%)
Query: 6 VKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVL--VHGHK 63
+++ S PF++R LK KG+ +E + ++ NK E NP +PVL GH
Sbjct: 24 IRVYSMRFCPFAQRTLMVLKAKGIRHEIININLKNKPEWFFEKNPFGL-VPVLENTQGH- 81
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARF-WANFTEQKLLETVFVAMCITGDEQE 122
I ES I EY+DE + + L P DPYE+A + + F++ L T F+ ++
Sbjct: 82 LITESVITCEYLDEAYPEKKLFPDDPYEKACQKMTFELFSKVPSLVTSFI-RAKRKEDHP 140
Query: 123 KALNEAREAMEKIEEVIEGKR--FFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLK 180
E ++ K+EE + KR FFGG ++ +D + W W E + ID
Sbjct: 141 GIKEELKKEFSKLEEAMANKRTAFFGGNSLSMIDYLI-W--PWFQRLEALELNECID--H 195
Query: 181 FPATTAWMTNFLSHPV 196
P WM PV
Sbjct: 196 TPKLKLWMATMQEDPV 211
>GTZ1_DIACA (P28342) Glutathione S-transferase 1 (EC 2.5.1.18) (SR8)
(GST class-zeta)
Length = 221
Score = 73.6 bits (179), Expect = 4e-13
Identities = 54/165 (32%), Positives = 88/165 (52%), Gaps = 13/165 (7%)
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSL---LLELNPVHKKIPVLV 59
+Q ++L SF S + RV AL LKG+++EY D+F L L+LNP+ +PVLV
Sbjct: 6 TQKMQLYSFSLSSCAWRVRIALHLKGLDFEYKAVDLFKGEHLTPEFLKLNPLGY-VPVLV 64
Query: 60 HGHKTIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTE------QKLLETVFVA 113
HG IA+S II Y++E + + PLLP D +RAL AN Q L ++
Sbjct: 65 HGDIVIADSLAIIMYLEEKFPENPLLPQDLQKRALNYQAANIVTSNIQPLQNLAVLNYIE 124
Query: 114 MCITGDEQ-EKALNEAREAMEKIEEVIEGK--RFFGGENIGYLDI 155
+ DE+ A + ++ +E++++G ++ G+ +G D+
Sbjct: 125 EKLGSDEKLSWAKHHIKKGFSALEKLLKGHAGKYATGDEVGLADL 169
>GSTZ_EUPES (P57108) Glutathione S-transferase zeta class (EC
2.5.1.18)
Length = 225
Score = 70.9 bits (172), Expect = 2e-12
Identities = 49/163 (30%), Positives = 88/163 (53%), Gaps = 15/163 (9%)
Query: 6 VKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSL---LLELNPVHKKIPVLVHGH 62
+KL S++ S S RV AL LKG++YEYV ++ L++NP+ +P LV G
Sbjct: 12 LKLYSYFRSSCSFRVRIALNLKGLDYEYVPVNLLKGEQFTPEFLKINPIGY-VPALVDGE 70
Query: 63 KTIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTE------QKLLETVFVAMCI 116
I++SF I+ Y++E + ++P+LP D +++A+ AN Q L F+ +
Sbjct: 71 DVISDSFAILMYLEEKYPEHPILPADIHKKAINYQAANIVSSSIQPLQNLAVLNFIGEKV 130
Query: 117 TGDEQ----EKALNEAREAMEKIEEVIEGKRFFGGENIGYLDI 155
+ DE+ ++ +++ A+EK+ + G RF G+ + D+
Sbjct: 131 SPDEKVPWVQRHISKGFAALEKLLQGHAG-RFATGDEVYLADL 172
>GTO1_HUMAN (P78417) Glutathione transferase omega 1 (EC 2.5.1.18)
(GSTO 1-1)
Length = 241
Score = 69.7 bits (169), Expect = 5e-12
Identities = 57/200 (28%), Positives = 90/200 (44%), Gaps = 11/200 (5%)
Query: 6 VKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH-KT 64
+++ S PF++R LK KG+ +E + ++ NK + NP +PVL + +
Sbjct: 24 IRIYSMRFCPFAERTRLVLKAKGIRHEVININLKNKPEWFFKKNPFGL-VPVLENSQGQL 82
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWAN-FTEQKLLETVFVAMCITGDEQEK 123
I ES I EY+DE + LLP DPYE+A + F++ L F+ ++
Sbjct: 83 IYESAITCEYLDEAYPGKKLLPDDPYEKACQKMILELFSKVPSLVGSFI-RSQNKEDYAG 141
Query: 124 ALNEAREAMEKIEEVIEGKR--FFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
E R+ K+EEV+ K+ FFGG +I +D + W W E + +D
Sbjct: 142 LKEEFRKEFTKLEEVLTNKKTTFFGGNSISMIDYLI-W--PWFERLEAMKLNECVD--HT 196
Query: 182 PATTAWMTNFLSHPVIKDTL 201
P WM P + L
Sbjct: 197 PKLKLWMAAMKEDPTVSALL 216
>GTZ1_ARATH (Q9ZVQ3) Glutathione S-transferase zeta-class 1 (EC
2.5.1.18) (AtGSTZ1) (Maleylacetone isomerase) (EC
5.2.1.-) (MAI)
Length = 221
Score = 68.6 bits (166), Expect = 1e-11
Identities = 37/96 (38%), Positives = 57/96 (58%), Gaps = 4/96 (4%)
Query: 2 GSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSL---LLELNPVHKKIPVL 58
G + +KL S+W S + RV AL LKG++YEY+ ++ ++NP+ +P L
Sbjct: 5 GEEKLKLYSYWRSSCAHRVRIALALKGLDYEYIPVNLLKGDQFDSDFKKINPMGT-VPAL 63
Query: 59 VHGHKTIAESFIIIEYIDETWKQYPLLPHDPYERAL 94
V G I +SF II Y+DE + + PLLP D ++RA+
Sbjct: 64 VDGDVVINDSFAIIMYLDEKYPEPPLLPRDLHKRAV 99
>GTO1_PIG (Q9N1F5) Glutathione transferase omega 1 (EC 2.5.1.18)
(GSTO 1-1) (Glutathione-dependent dehydroascorbate
reductase)
Length = 241
Score = 65.5 bits (158), Expect = 1e-10
Identities = 55/206 (26%), Positives = 93/206 (44%), Gaps = 14/206 (6%)
Query: 6 VKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH-KT 64
+++ S PF++R L KG+ ++ + ++ NK + NP +PVL + +
Sbjct: 24 IRVYSMRFCPFAQRTLLVLNAKGIRHQVININLKNKPEWFFQKNP-SGLVPVLENSQGQL 82
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
I ES I EY+DE + LLP DPYE+A + F + + + +E + +
Sbjct: 83 IYESAITCEYLDEAYPGKKLLPDDPYEKACQKM--VFELSSKVPPLLIRFIRRENEADCS 140
Query: 125 --LNEAREAMEKIEEVIEGKR--FFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLK 180
E R+ K+EEV+ K+ +FGG ++ +D + W W E + ID
Sbjct: 141 GLKEELRKEFSKLEEVLTKKKTTYFGGSSLSMIDYLI-W--PWFERLEALELNECID--H 195
Query: 181 FPATTAWMTNFLSHPVIKDT-LPPRD 205
P WM + P + + PRD
Sbjct: 196 TPKLKLWMAAMMKDPAVSALHIEPRD 221
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,747,059
Number of Sequences: 164201
Number of extensions: 1283648
Number of successful extensions: 3574
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 3483
Number of HSP's gapped (non-prelim): 84
length of query: 228
length of database: 59,974,054
effective HSP length: 106
effective length of query: 122
effective length of database: 42,568,748
effective search space: 5193387256
effective search space used: 5193387256
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0162b.4


