

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
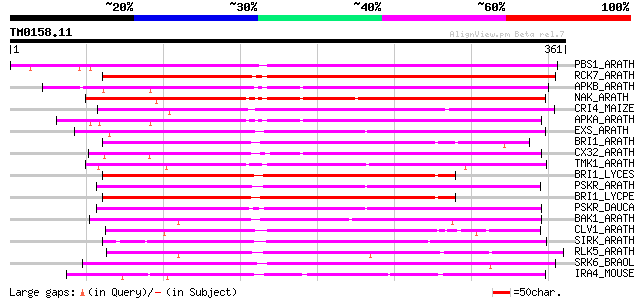
Query= TM0158.11
(361 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 266 8e-71
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 245 1e-64
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 223 8e-58
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 222 1e-57
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 221 3e-57
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 220 4e-57
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 207 3e-53
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 207 3e-53
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 205 2e-52
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 204 2e-52
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 201 2e-51
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 201 3e-51
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 201 3e-51
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 200 4e-51
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 192 1e-48
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 189 7e-48
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 189 1e-47
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 187 5e-47
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 185 1e-46
IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (... 174 2e-43
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 266 bits (679), Expect = 8e-71
Identities = 161/376 (42%), Positives = 216/376 (56%), Gaps = 25/376 (6%)
Query: 1 MSCFNCCEEDDF----------HKTAESGGPYVVKNPSGNDGNHHASETAKQGG---QTV 47
M CF+C + D H + P V N SG + GG + +
Sbjct: 1 MGCFSCFDSSDDEKLNPVDESNHGQKKQSQPTVSNNISGLPSGGEKLSSKTNGGSKRELL 60
Query: 48 KPQP----IEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKN-GQAAAIKKLDAS 102
P+ I + EL T NF D+ +GEG +GRVY G L + GQ A+K+LD +
Sbjct: 61 LPRDGLGQIAAHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRN 120
Query: 103 K-QPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKG 161
Q + EFL +V M+S L H N V L+GY DG+ R+LVYEF GSL D LH K
Sbjct: 121 GLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKE 180
Query: 162 AQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLS 221
A L W R+KIA GAA+GLE+LH+KA+P +I+RD KSSN+L+ + K++DF L+
Sbjct: 181 A-----LDWNMRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLA 235
Query: 222 NQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLP 281
P STRV+GT+GY APEYAMTGQL KSDVYSFGVV LEL+TGRK +D +P
Sbjct: 236 KLGPTGDKSHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMP 295
Query: 282 RGQQSLVTWATPKLSE-DKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSI 340
G+Q+LV WA P ++ K + D RL G +P +A+ + AVA++C+Q +A RP ++
Sbjct: 296 HGEQNLVAWARPLFNDRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIAD 355
Query: 341 VVKALQPLLTARPGPA 356
VV AL L P+
Sbjct: 356 VVTALSYLANQAYDPS 371
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 245 bits (625), Expect = 1e-64
Identities = 137/298 (45%), Positives = 190/298 (62%), Gaps = 8/298 (2%)
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVL-KNGQAAAIKKLDASK-QPDEEFLAQVSMVSR 118
EL E T NF D +GEG +G+V+ G + K Q AIK+LD + Q EF+ +V +S
Sbjct: 95 ELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVLTLSL 154
Query: 119 LKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAV 178
H N V+L+G+ +G+ R+LVYE+ GSL D LH G +P L W R+KIA
Sbjct: 155 ADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPS--GKKP---LDWNTRMKIAA 209
Query: 179 GAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLG 238
GAARGLEYLH++ P +I+RD+K SN+L+ +D K++DF L+ P STRV+G
Sbjct: 210 GAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTRVMG 269
Query: 239 TFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSED 298
T+GY AP+YAMTGQL KSD+YSFGVVLLEL+TGRK +D+T R Q+LV WA P +
Sbjct: 270 TYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFKDR 329
Query: 299 K-VRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGP 355
+ + VD L G+YP + + + A++A+CVQ + RP +S VV AL L +++ P
Sbjct: 330 RNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNFLASSKYDP 387
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 223 bits (567), Expect = 8e-58
Identities = 133/344 (38%), Positives = 203/344 (58%), Gaps = 22/344 (6%)
Query: 22 YVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISE---DELKEVTDNFGQDSLIGEG 78
Y+ + + G+ +S + + +T + + ++ PN+ ELK T NF DS++GEG
Sbjct: 20 YMSSEANDSLGSKSSSVSIRTNPRT-EGEILQSPNLKSFTFAELKAATRNFRPDSVLGEG 78
Query: 79 SYGRVYYGVLKN----------GQAAAIKKLDASK-QPDEEFLAQVSMVSRLKHENFVQL 127
+G V+ G + G A+KKL+ Q +E+LA+V+ + + H N V+L
Sbjct: 79 GFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQGHQEWLAEVNYLGQFSHPNLVKL 138
Query: 128 LGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYL 187
+GY ++ R+LVYEF GSL + L R QP L+W R+K+A+GAA+GL +L
Sbjct: 139 IGYCLEDEHRLLVYEFMPRGSLENHLFRRGSY--FQP---LSWTLRLKVALGAAKGLAFL 193
Query: 188 HEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEY 247
H A+ +I+RD K+SN+L+ + AK++DF L+ P STR++GT+GY APEY
Sbjct: 194 HN-AETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKSHVSTRIMGTYGYAAPEY 252
Query: 248 AMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKL-SEDKVRQCVDT 306
TG L KSDVYS+GVVLLE+L+GR+ VD P G+Q LV WA P L ++ K+ + +D
Sbjct: 253 LATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVEWARPLLANKRKLFRVIDN 312
Query: 307 RLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLT 350
RL +Y + K+A +A C+ +E RPNM+ VV L+ + T
Sbjct: 313 RLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHLEHIQT 356
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 222 bits (565), Expect = 1e-57
Identities = 133/312 (42%), Positives = 192/312 (60%), Gaps = 20/312 (6%)
Query: 50 QPIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAA----------AIKKL 99
Q + N S ELK T NF DS++GEG +G V+ G + A A+K+L
Sbjct: 49 QNANLKNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRL 108
Query: 100 DASK-QPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKG 158
+ Q E+LA+++ + +L H N V+L+GY ++ R+LVYEF + GSL + L R+G
Sbjct: 109 NQEGFQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLF-RRG 167
Query: 159 VKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADF 218
QP L+W RV++A+GAARGL +LH A P +I+RD K+SN+L+ + AK++DF
Sbjct: 168 TF-YQP---LSWNTRVRMALGAARGLAFLHN-AQPQVIYRDFKASNILLDSNYNAKLSDF 222
Query: 219 DLSNQAPDMAARLH-STRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVD 277
L+ P M H STRV+GT GY APEY TG L+ KSDVYSFGVVLLELL+GR+ +D
Sbjct: 223 GLARDGP-MGDNSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAID 281
Query: 278 HTLPRGQQSLVTWATPKL-SEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRP 336
P G+ +LV WA P L ++ ++ + +D RL G+Y K+A +A C+ +A RP
Sbjct: 282 KNQPVGEHNLVDWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRP 341
Query: 337 NMSIVVKALQPL 348
M+ +VK ++ L
Sbjct: 342 TMNEIVKTMEEL 353
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 221 bits (562), Expect = 3e-57
Identities = 130/300 (43%), Positives = 180/300 (59%), Gaps = 9/300 (3%)
Query: 58 SEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKK-LDAS--KQPDEEFLAQVS 114
S +EL++ T F +DS +G+GS+ V+ G+L++G A+K+ + AS K+ +EF ++
Sbjct: 494 SYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNELD 553
Query: 115 MVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRV 174
++SRL H + + LLGY DG+ R+LVYEF ++GSL+ LHG K L WA+RV
Sbjct: 554 LLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHG----KDPNLKKRLNWARRV 609
Query: 175 KIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHST 234
IAV AARG+EYLH A P +IHRDIKSSN+LI +D A++ADF LS P + S
Sbjct: 610 TIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSE 669
Query: 235 RVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPK 294
GT GY PEY L KSDVYSFGVVLLE+L+GRK +D G ++V WA P
Sbjct: 670 LPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEEG--NIVEWAVPL 727
Query: 295 LSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLLTARPG 354
+ + +D L +A+ K+A+VA CV+ RP+M V AL+ L G
Sbjct: 728 IKAGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHALALLMG 787
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 220 bits (561), Expect = 4e-57
Identities = 136/337 (40%), Positives = 199/337 (58%), Gaps = 27/337 (8%)
Query: 31 DGNHHASETAKQGGQTVKPQP------IEVPNI---SEDELKEVTDNFGQDSLIGEGSYG 81
D S +K +V+P P ++ PN+ S ELK T NF DS++GEG +G
Sbjct: 21 DAKDIGSLGSKASSVSVRPSPRTEGEILQSPNLKSFSFAELKSATRNFRPDSVLGEGGFG 80
Query: 82 RVYYGVLKN----------GQAAAIKKLDASK-QPDEEFLAQVSMVSRLKHENFVQLLGY 130
V+ G + G A+KKL+ Q +E+LA+V+ + + H + V+L+GY
Sbjct: 81 CVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQGHQEWLAEVNYLGQFSHRHLVKLIGY 140
Query: 131 SVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEK 190
++ R+LVYEF GSL + L R+G+ QP L+W R+K+A+GAA+GL +LH
Sbjct: 141 CLEDEHRLLVYEFMPRGSLENHLF-RRGLY-FQP---LSWKLRLKVALGAAKGLAFLHS- 194
Query: 191 ADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMT 250
++ +I+RD K+SN+L+ + AK++DF L+ P STRV+GT GY APEY T
Sbjct: 195 SETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVSTRVMGTHGYAAPEYLAT 254
Query: 251 GQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKL-SEDKVRQCVDTRLG 309
G L KSDVYSFGVVLLELL+GR+ VD P G+++LV WA P L ++ K+ + +D RL
Sbjct: 255 GHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKPYLVNKRKIFRVIDNRLQ 314
Query: 310 GEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
+Y + K+A ++ C+ E RPNMS VV L+
Sbjct: 315 DQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLE 351
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 207 bits (528), Expect = 3e-53
Identities = 121/315 (38%), Positives = 179/315 (56%), Gaps = 15/315 (4%)
Query: 43 GGQTVKPQPIEVPNISEDELK-------EVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAA 95
G ++ +P I + + LK E TD+F + ++IG+G +G VY L + A
Sbjct: 884 GSRSREPLSINIAMFEQPLLKVRLGDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVA 943
Query: 96 IKKLDASK-QPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILH 154
+KKL +K Q + EF+A++ + ++KH N V LLGY ++LVYE+ NGSL L
Sbjct: 944 VKKLSEAKTQGNREFMAEMETLGKVKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLR 1003
Query: 155 GRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAK 214
+ G+ VL W++R+KIAVGAARGL +LH PHIIHRDIK+SN+L+ D K
Sbjct: 1004 NQTGML-----EVLDWSKRLKIAVGAARGLAFLHHGFIPHIIHRDIKASNILLDGDFEPK 1058
Query: 215 IADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRK 274
+ADF L+ + + ST + GTFGY PEY + + K DVYSFGV+LLEL+TG++
Sbjct: 1059 VADFGLARLISACESHV-STVIAGTFGYIPPEYGQSARATTKGDVYSFGVILLELVTGKE 1117
Query: 275 PVDHTLPRGQ-QSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEAD 333
P + +LV WA K+++ K +D L + ++ +A LC+
Sbjct: 1118 PTGPDFKESEGGNLVGWAIQKINQGKAVDVIDPLLVSVALKNSQLRLLQIAMLCLAETPA 1177
Query: 334 FRPNMSIVVKALQPL 348
RPNM V+KAL+ +
Sbjct: 1178 KRPNMLDVLKALKEI 1192
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 207 bits (528), Expect = 3e-53
Identities = 120/281 (42%), Positives = 167/281 (58%), Gaps = 10/281 (3%)
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKL-DASKQPDEEFLAQVSMVSRL 119
+L + T+ F DSLIG G +G VY +LK+G A AIKKL S Q D EF+A++ + ++
Sbjct: 875 DLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEMETIGKI 934
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
KH N V LLGY G+ R+LVYEF GSL D+LH K + G L W+ R KIA+G
Sbjct: 935 KHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPK-----KAGVKLNWSTRRKIAIG 989
Query: 180 AARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
+ARGL +LH PHIIHRD+KSSNVL+ ++ A+++DF ++ M L + + GT
Sbjct: 990 SARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGT 1049
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDK 299
GY PEY + + + K DVYS+GVVLLELLTG++P D + G +LV W + ++ +
Sbjct: 1050 PGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD-SPDFGDNNLVGW-VKQHAKLR 1107
Query: 300 VRQCVDTRLGGEYPPKAVAKM--AAVAALCVQYEADFRPNM 338
+ D L E P + + VA C+ A RP M
Sbjct: 1108 ISDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTM 1148
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 205 bits (521), Expect = 2e-52
Identities = 122/310 (39%), Positives = 180/310 (57%), Gaps = 24/310 (7%)
Query: 52 IEVPNISED---ELKEVTDNFGQDSLIGEGSYGRVYYGVLK----------NGQAAAIKK 98
+E PN+ +LK T NF DS++G+G +G+VY G + +G AIK+
Sbjct: 66 LESPNLKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKR 125
Query: 99 LDA-SKQPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRK 157
L++ S Q E+ ++V+ + L H N V+LLGY + +LVYEF GSL L R
Sbjct: 126 LNSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFRRN 185
Query: 158 GVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIAD 217
P P W R+KI +GAARGL +LH +I+RD K+SN+L+ + AK++D
Sbjct: 186 -----DPFP---WDLRIKIVIGAARGLAFLHS-LQREVIYRDFKASNILLDSNYDAKLSD 236
Query: 218 FDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVD 277
F L+ P +TR++GT+GY APEY TG L KSDV++FGVVLLE++TG +
Sbjct: 237 FGLAKLGPADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHN 296
Query: 278 HTLPRGQQSLVTWATPKLS-EDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRP 336
PRGQ+SLV W P+LS + +V+Q +D + G+Y K +MA + C++ + RP
Sbjct: 297 TKRPRGQESLVDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRP 356
Query: 337 NMSIVVKALQ 346
+M VV+ L+
Sbjct: 357 HMKEVVEVLE 366
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 204 bits (520), Expect = 2e-52
Identities = 121/308 (39%), Positives = 182/308 (58%), Gaps = 13/308 (4%)
Query: 50 QPIEVPN--ISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLD---ASKQ 104
Q +E N IS L+ VT+NF D+++G G +G VY G L +G A+K+++ + +
Sbjct: 567 QMVEAGNMLISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGK 626
Query: 105 PDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQP 164
EF ++++++++++H + V LLGY +DGN ++LVYE+ G+L L +G +P
Sbjct: 627 GFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLF-EWSEEGLKP 685
Query: 165 GPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQA 224
L W QR+ +A+ ARG+EYLH A IHRD+K SN+L+ DD AK+ADF L A
Sbjct: 686 ---LLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLA 742
Query: 225 PDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQ 284
P+ + TR+ GTFGY APEYA+TG++ K DVYSFGV+L+EL+TGRK +D + P
Sbjct: 743 PEGKGSIE-TRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEES 801
Query: 285 QSLVTWATPKL--SEDKVRQCVDTRLG-GEYPPKAVAKMAAVAALCVQYEADFRPNMSIV 341
LV+W E ++ +DT + E +V +A +A C E RP+M
Sbjct: 802 IHLVSWFKRMYINKEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHA 861
Query: 342 VKALQPLL 349
V L L+
Sbjct: 862 VNILSSLV 869
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 201 bits (511), Expect = 2e-51
Identities = 109/231 (47%), Positives = 144/231 (62%), Gaps = 7/231 (3%)
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKL-DASKQPDEEFLAQVSMVSRL 119
+L E T+ F DSL+G G +G VY LK+G AIKKL S Q D EF A++ + ++
Sbjct: 880 DLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETIGKI 939
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
KH N V LLGY G R+LVYE+ GSL D+LH RK + G L W R KIA+G
Sbjct: 940 KHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKI-----GIKLNWPARRKIAIG 994
Query: 180 AARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
AARGL +LH PHIIHRD+KSSNVL+ ++ A+++DF ++ M L + + GT
Sbjct: 995 AARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGT 1054
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTW 290
GY PEY + + + K DVYS+GVVLLELLTG++P D + G +LV W
Sbjct: 1055 PGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGW 1104
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 201 bits (510), Expect = 3e-51
Identities = 116/291 (39%), Positives = 165/291 (55%), Gaps = 9/291 (3%)
Query: 57 ISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDAS-KQPDEEFLAQVSM 115
+S D+L + T++F Q ++IG G +G VY L +G+ AIKKL Q + EF A+V
Sbjct: 722 LSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVET 781
Query: 116 VSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGP-VLTWAQRV 174
+SR +H N V L G+ N R+L+Y + NGSL LH R GP +L W R+
Sbjct: 782 LSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERND------GPALLKWKTRL 835
Query: 175 KIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHST 234
+IA GAA+GL YLHE DPHI+HRDIKSSN+L+ ++ + +ADF L+ + ST
Sbjct: 836 RIAQGAAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHV-ST 894
Query: 235 RVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPK 294
++GT GY PEY K DVYSFGVVLLELLT ++PVD P+G + L++W
Sbjct: 895 DLVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKM 954
Query: 295 LSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKAL 345
E + + D + + K + ++ +A LC+ RP +V L
Sbjct: 955 KHESRASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWL 1005
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 201 bits (510), Expect = 3e-51
Identities = 109/231 (47%), Positives = 144/231 (62%), Gaps = 7/231 (3%)
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKL-DASKQPDEEFLAQVSMVSRL 119
+L E T+ F DSL+G G +G VY LK+G AIKKL S Q D EF A++ + ++
Sbjct: 880 DLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETIGKI 939
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
KH N V LLGY G R+LVYE+ GSL D+LH RK + G L W R KIA+G
Sbjct: 940 KHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRK-----KTGIKLNWPARRKIAIG 994
Query: 180 AARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
AARGL +LH PHIIHRD+KSSNVL+ ++ A+++DF ++ M L + + GT
Sbjct: 995 AARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGT 1054
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTW 290
GY PEY + + + K DVYS+GVVLLELLTG++P D + G +LV W
Sbjct: 1055 PGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGW 1104
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 200 bits (509), Expect = 4e-51
Identities = 116/291 (39%), Positives = 164/291 (55%), Gaps = 7/291 (2%)
Query: 57 ISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASK-QPDEEFLAQVSM 115
+S D++ + T +F Q ++IG G +G VY L +G AIK+L Q D EF A+V
Sbjct: 731 LSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVET 790
Query: 116 VSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVK 175
+SR +H N V LLGY N ++L+Y + NGSL LH + V G P L W R++
Sbjct: 791 LSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEK--VDGP---PSLDWKTRLR 845
Query: 176 IAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTR 235
IA GAA GL YLH+ +PHI+HRDIKSSN+L+ D VA +ADF L+ + +T
Sbjct: 846 IARGAAEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHV-TTD 904
Query: 236 VLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKL 295
++GT GY PEY K DVYSFGVVLLELLTGR+P+D PRG + L++W
Sbjct: 905 LVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMK 964
Query: 296 SEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
+E + + D + + + + + +A C+ RP +V L+
Sbjct: 965 TEKRESEIFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLE 1015
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 192 bits (487), Expect = 1e-48
Identities = 110/298 (36%), Positives = 167/298 (55%), Gaps = 10/298 (3%)
Query: 53 EVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEE--FL 110
++ S EL+ +DNF +++G G +G+VY G L +G A+K+L + E F
Sbjct: 273 QLKRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQ 332
Query: 111 AQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTW 170
+V M+S H N ++L G+ + R+LVY + +NGS+ L R + P L W
Sbjct: 333 TEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERP-----ESQPPLDW 387
Query: 171 AQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAAR 230
+R +IA+G+ARGL YLH+ DP IIHRD+K++N+L+ ++ A + DF L+ + D
Sbjct: 388 PKRQRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLA-KLMDYKDT 446
Query: 231 LHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQS--LV 288
+T V GT G+ APEY TG+ + K+DV+ +GV+LLEL+TG++ D L+
Sbjct: 447 HVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLL 506
Query: 289 TWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
W L E K+ VD L G Y + V ++ VA LC Q RP MS VV+ L+
Sbjct: 507 DWVKGLLKEKKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLE 564
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 189 bits (481), Expect = 7e-48
Identities = 116/294 (39%), Positives = 173/294 (58%), Gaps = 23/294 (7%)
Query: 63 KEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKL--DASKQPDEEFLAQVSMVSRLK 120
++V + ++++IG+G G VY G + N AIK+L + + D F A++ + R++
Sbjct: 686 EDVLECLKEENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIR 745
Query: 121 HENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGA 180
H + V+LLGY + ++ +L+YE+ NGSL ++LHG KG L W R ++AV A
Sbjct: 746 HRHIVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSKGGH-------LQWETRHRVAVEA 798
Query: 181 ARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTF 240
A+GL YLH P I+HRD+KS+N+L+ D A +ADF L+ D AA + + G++
Sbjct: 799 AKGLCYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSY 858
Query: 241 GYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKV 300
GY APEYA T +++ KSDVYSFGVVLLEL+ G+KPV G +V W + +E+++
Sbjct: 859 GYIAPEYAYTLKVDEKSDVYSFGVVLLELIAGKKPVGE-FGEG-VDIVRWV--RNTEEEI 914
Query: 301 RQ---------CVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKAL 345
Q VD RL G YP +V + +A +CV+ EA RP M VV L
Sbjct: 915 TQPSDAAIVVAIVDPRLTG-YPLTSVIHVFKIAMMCVEEEAAARPTMREVVHML 967
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 189 bits (479), Expect = 1e-47
Identities = 111/290 (38%), Positives = 172/290 (59%), Gaps = 12/290 (4%)
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKL-DASKQPDEEFLAQVSMVSRL 119
E+ +T+NF + +IG+G +G+VY+GV+ NG+ A+K L + S Q +EF A+V ++ R+
Sbjct: 568 EVVNITNNF--ERVIGKGGFGKVYHGVI-NGEQVAVKVLSEESAQGYKEFRAEVDLLMRV 624
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
H N L+GY + N +L+YE+ +N +L D L G++ +L+W +R+KI++
Sbjct: 625 HHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRSF-------ILSWEERLKISLD 677
Query: 180 AARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
AA+GLEYLH P I+HRD+K +N+L+ + AK+ADF LS + ST V G+
Sbjct: 678 AAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGS 737
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDK 299
GY PEY T Q+N KSDVYS GVVLLE++TG +P + + + L+
Sbjct: 738 IGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITG-QPAIASSKTEKVHISDHVRSILANGD 796
Query: 300 VRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLL 349
+R VD RL Y + KM+ +A C ++ + RP MS VV L+ ++
Sbjct: 797 IRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMELKQIV 846
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 187 bits (474), Expect = 5e-47
Identities = 116/311 (37%), Positives = 177/311 (56%), Gaps = 23/311 (7%)
Query: 64 EVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDAS-KQPDEE----------FLAQ 112
E+ D + ++IG GS G+VY L+ G+ A+KKL+ S K D+E F A+
Sbjct: 678 EIADCLDEKNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAE 737
Query: 113 VSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHG-RKGVKGAQPGPVLTWA 171
V + ++H++ V+L G+ ++LVYE+ NGSL D+LHG RKG G VL W
Sbjct: 738 VETLGTIRHKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKG------GVVLGWP 791
Query: 172 QRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARL 231
+R++IA+ AA GL YLH P I+HRD+KSSN+L+ D AK+ADF ++ ++
Sbjct: 792 ERLRIALDAAEGLSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKT 851
Query: 232 HS--TRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVT 289
+ + G+ GY APEY T ++N KSD+YSFGVVLLEL+TG++P D L G + +
Sbjct: 852 PEAMSGIAGSCGYIAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTDSEL--GDKDMAK 909
Query: 290 WATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLL 349
W L + + +D +L ++ + ++K+ + LC RP+M VV LQ +
Sbjct: 910 WVCTALDKCGLEPVIDPKLDLKF-KEEISKVIHIGLLCTSPLPLNRPSMRKVVIMLQEVS 968
Query: 350 TARPGPAGETA 360
A P + T+
Sbjct: 969 GAVPCSSPNTS 979
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 185 bits (470), Expect = 1e-46
Identities = 109/316 (34%), Positives = 178/316 (55%), Gaps = 14/316 (4%)
Query: 48 KPQPIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLD-ASKQPD 106
K + +E+P I + + + T+NF + +G+G +G VY G L +G+ A+K+L S Q
Sbjct: 507 KFEELELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGT 566
Query: 107 EEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGP 166
+EF+ +V++++RL+H N VQ+LG ++G+ ++L+YE+ N SL L G+
Sbjct: 567 DEFMNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSK----- 621
Query: 167 VLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPD 226
L W +R I G ARGL YLH+ + IIHRD+K SN+L+ + + KI+DF ++
Sbjct: 622 -LNWNERFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFER 680
Query: 227 MAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQS 286
++ +V+GT+GY +PEYAM G + KSDV+SFGV++LE+++G+K +
Sbjct: 681 DETEANTMKVVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYEND 740
Query: 287 LVTWATPKLSEDKVRQCVDTRLGGE-------YPPKAVAKMAAVAALCVQYEADFRPNMS 339
L+++ + E + + VD + + P+ V K + LCVQ A+ RP MS
Sbjct: 741 LLSYVWSRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMS 800
Query: 340 IVVKALQPLLTARPGP 355
VV T P P
Sbjct: 801 SVVWMFGSEATEIPQP 816
>IRA4_MOUSE (Q8R4K2) Interleukin-1 receptor-associated kinase-4 (EC
2.7.1.37) (IRAK-4)
Length = 459
Score = 174 bits (442), Expect = 2e-43
Identities = 112/322 (34%), Positives = 179/322 (54%), Gaps = 24/322 (7%)
Query: 38 ETAKQGGQTVKPQPIEVPNISEDELKEVTDNFGQD------SLIGEGSYGRVYYGVLKNG 91
+++ ++V+ + S ELK +T+NF + + +GEG +G VY G + N
Sbjct: 149 DSSSPDNRSVESSDTRFHSFSFHELKSITNNFDEQPASAGGNRMGEGGFGVVYKGCVNN- 207
Query: 92 QAAAIKKLDA-----SKQPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASN 146
A+KKL A +++ ++F ++ +++ +HEN V+LLG+S D ++ LVY + N
Sbjct: 208 TIVAVKKLGAMVEISTEELKQQFDQEIKVMATCQHENLVELLGFSSDSDNLCLVYAYMPN 267
Query: 147 GSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVL 206
GSL D L G P L+W R K+A G A G+ +LHE H IHRDIKS+N+L
Sbjct: 268 GSLLDRLSCLDGT------PPLSWHTRCKVAQGTANGIRFLHEN---HHIHRDIKSANIL 318
Query: 207 IFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVL 266
+ D AKI+DF L+ + +A + ++R++GT Y APE A+ G++ KSD+YSFGVVL
Sbjct: 319 LDKDFTAKISDFGLARASARLAQTVMTSRIVGTTAYMAPE-ALRGEITPKSDIYSFGVVL 377
Query: 267 LELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAAL 326
LEL+TG VD R Q L+ E+K + + P +V M + A+
Sbjct: 378 LELITGLAAVDEN--REPQLLLDIKEEIEDEEKTIEDYTDEKMSDADPASVEAMYSAASQ 435
Query: 327 CVQYEADFRPNMSIVVKALQPL 348
C+ + + RP+++ V + LQ +
Sbjct: 436 CLHEKKNRRPDIAKVQQLLQEM 457
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,531,617
Number of Sequences: 164201
Number of extensions: 1987231
Number of successful extensions: 9560
Number of sequences better than 10.0: 1636
Number of HSP's better than 10.0 without gapping: 954
Number of HSP's successfully gapped in prelim test: 687
Number of HSP's that attempted gapping in prelim test: 6027
Number of HSP's gapped (non-prelim): 1860
length of query: 361
length of database: 59,974,054
effective HSP length: 111
effective length of query: 250
effective length of database: 41,747,743
effective search space: 10436935750
effective search space used: 10436935750
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0158.11


