

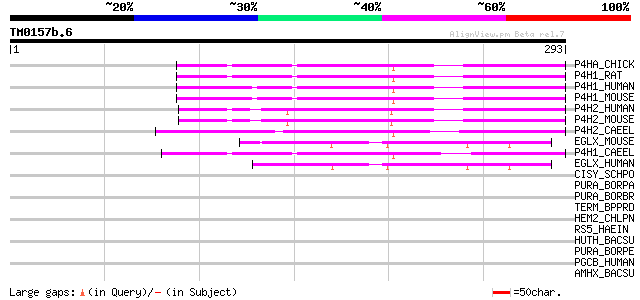
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0157b.6
(293 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
P4HA_CHICK (P16924) Prolyl 4-hydroxylase alpha subunit (EC 1.14.... 94 4e-19
P4H1_RAT (P54001) Prolyl 4-hydroxylase alpha-1 subunit precursor... 90 8e-18
P4H1_HUMAN (P13674) Prolyl 4-hydroxylase alpha-1 subunit precurs... 87 4e-17
P4H1_MOUSE (Q60715) Prolyl 4-hydroxylase alpha-1 subunit precurs... 87 5e-17
P4H2_HUMAN (O15460) Prolyl 4-hydroxylase alpha-2 subunit precurs... 81 3e-15
P4H2_MOUSE (Q60716) Prolyl 4-hydroxylase alpha-2 subunit precurs... 79 1e-14
P4H2_CAEEL (Q20065) Prolyl 4-hydroxylase alpha-2 subunit precurs... 79 1e-14
EGLX_MOUSE (Q8BG58) Putative HIF-prolyl hydroxylase PH-4 (EC 1.1... 71 3e-12
P4H1_CAEEL (Q10576) Prolyl 4-hydroxylase alpha-1 subunit precurs... 69 1e-11
EGLX_HUMAN (Q9NXG6) Putative HIF-prolyl hydroxylase PH-4 (EC 1.1... 69 1e-11
CISY_SCHPO (Q10306) Probable citrate synthase, mitochondrial pre... 33 1.1
PURA_BORPA (Q7W6Q7) Adenylosuccinate synthetase (EC 6.3.4.4) (IM... 32 1.9
PURA_BORBR (Q7WHP1) Adenylosuccinate synthetase (EC 6.3.4.4) (IM... 32 1.9
TERM_BPPRD (P09009) DNA terminal protein (Protein P8) 32 2.4
HEM2_CHLPN (Q9Z7G1) Delta-aminolevulinic acid dehydratase (EC 4.... 32 2.4
RS5_HAEIN (P44374) 30S ribosomal protein S5 31 3.2
HUTH_BACSU (P10944) Histidine ammonia-lyase (EC 4.3.1.3) (Histid... 30 5.4
PURA_BORPE (Q7VWM1) Adenylosuccinate synthetase (EC 6.3.4.4) (IM... 30 7.1
PGCB_HUMAN (Q96GW7) Brevican core protein precursor (Brain enric... 30 7.1
AMHX_BACSU (P54983) Amidohydrolase amhX (EC 3.5.1.-) (Aminoacylase) 30 7.1
>P4HA_CHICK (P16924) Prolyl 4-hydroxylase alpha subunit (EC
1.14.11.2) (4-PH alpha)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase alpha
subunit)
Length = 516
Score = 94.0 bits (232), Expect = 4e-19
Identities = 61/210 (29%), Positives = 103/210 (49%), Gaps = 24/210 (11%)
Query: 89 KPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQ-NTKGIRTSSGVFVSSSED 147
KPR + F + + E+ +++ +AK L S + + ET + T R S ++S E
Sbjct: 316 KPRIVRFLDIISDEEIETVKELAKPRL--SRATVHDPETGKLTTAHYRVSKSAWLSGYES 373
Query: 148 KTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKS----QR 203
++ I +I T + S E + Y V +Y PH+D E K R
Sbjct: 374 PV--VSRINTRIQDLTGLDVSTAEELQVANYGVGGQYEPHFDFGRKDEPDAFKELGTGNR 431
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGTI 263
+A++L Y++DV GG T+FP +G V P++G + +Y+LFP+G
Sbjct: 432 IATWLFYMSDVSAGGATVFPE---------------VGASVWPKKGTAVFWYNLFPSGEG 476
Query: 264 DPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
D ++ H +CPV+ G KWV+ KW+ + Q++
Sbjct: 477 DYSTRHAACPVLVGNKWVSNKWLHERGQEF 506
>P4H1_RAT (P54001) Prolyl 4-hydroxylase alpha-1 subunit precursor
(EC 1.14.11.2) (4-PH alpha-1)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-1 subunit)
Length = 534
Score = 89.7 bits (221), Expect = 8e-18
Identities = 59/210 (28%), Positives = 101/210 (48%), Gaps = 24/210 (11%)
Query: 89 KPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQ-NTKGIRTSSGVFVSSSED 147
KPR + F + + + + + +AK L S + + ET + T R S ++S ED
Sbjct: 334 KPRIIRFHDIISDAEIEIVKDLAKPRL--SRATVHDPETGKLTTAQYRVSKSAWLSGYED 391
Query: 148 KTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKS----QR 203
++ I +I T + S E + Y V +Y PH+D E + R
Sbjct: 392 PV--VSRINMRIQDLTGLDVSTAEELQVANYGVGGQYEPHFDFARKDEPDAFRELGTGNR 449
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGTI 263
+A++L Y++DV GG T+FP +G V P++G + +Y+LF +G
Sbjct: 450 IATWLFYMSDVSAGGATVFPE---------------VGASVWPKKGTAVFWYNLFASGEG 494
Query: 264 DPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
D ++ H +CPV+ G KWV+ KW+ + Q++
Sbjct: 495 DYSTRHAACPVLVGNKWVSNKWLHERGQEF 524
>P4H1_HUMAN (P13674) Prolyl 4-hydroxylase alpha-1 subunit precursor
(EC 1.14.11.2) (4-PH alpha-1)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-1 subunit)
Length = 534
Score = 87.4 bits (215), Expect = 4e-17
Identities = 58/210 (27%), Positives = 102/210 (47%), Gaps = 24/210 (11%)
Query: 89 KPRALYFPNFATAEQCKSIVGVAKAGLKPSALALR-EGETEQNTKGIRTSSGVFVSSSED 147
KPR + F + + + + + +AK L+ + ++ G+ E T R S ++S E+
Sbjct: 334 KPRIIRFHDIISDAEIEIVKDLAKPRLRRATISNPITGDLE--TVHYRISKSAWLSGYEN 391
Query: 148 KTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKS----QR 203
++ I +I T + S E + Y V +Y PH+D E K R
Sbjct: 392 PV--VSRINMRIQDLTGLDVSTAEELQVANYGVGGQYEPHFDFARKDEPDAFKELGTGNR 449
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGTI 263
+A++L Y++DV GG T+FP +G V P++G + +Y+LF +G
Sbjct: 450 IATWLFYMSDVSAGGATVFPE---------------VGASVWPKKGTAVFWYNLFASGEG 494
Query: 264 DPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
D ++ H +CPV+ G KWV+ KW+ + Q++
Sbjct: 495 DYSTRHAACPVLVGNKWVSNKWLHERGQEF 524
>P4H1_MOUSE (Q60715) Prolyl 4-hydroxylase alpha-1 subunit precursor
(EC 1.14.11.2) (4-PH alpha-1)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-1 subunit)
Length = 534
Score = 87.0 bits (214), Expect = 5e-17
Identities = 58/210 (27%), Positives = 101/210 (47%), Gaps = 24/210 (11%)
Query: 89 KPRALYFPNFATAEQCKSIVGVAKAGLKPSALALR-EGETEQNTKGIRTSSGVFVSSSED 147
KPR + F + + + + + +AK L+ + ++ G E T R S ++S ED
Sbjct: 334 KPRIIRFHDIISDAEIEIVKDLAKPRLRRATISNPVTGALE--TVHYRISKSAWLSGYED 391
Query: 148 KTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKS----QR 203
++ I +I T + S E + Y V +Y PH+D E + R
Sbjct: 392 PV--VSRINMRIQDLTGLDVSTAEELQVANYGVGGQYEPHFDFARKDEPDAFRELGTGNR 449
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGTI 263
+A++L Y++DV GG T+FP +G V P++G + +Y+LF +G
Sbjct: 450 IATWLFYMSDVSAGGATVFPE---------------VGASVWPKKGTAVFWYNLFASGEG 494
Query: 264 DPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
D ++ H +CPV+ G KWV+ KW+ + Q++
Sbjct: 495 DYSTRHAACPVLVGNKWVSNKWLHERGQEF 524
>P4H2_HUMAN (O15460) Prolyl 4-hydroxylase alpha-2 subunit precursor
(EC 1.14.11.2) (4-PH alpha-2)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-2 subunit) (UNQ290/PRO330)
Length = 535
Score = 80.9 bits (198), Expect = 3e-15
Identities = 56/212 (26%), Positives = 98/212 (45%), Gaps = 30/212 (14%)
Query: 90 PRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS---- 145
P + + + + E+ + I +AK L + +R+ +T G+ T + VS S
Sbjct: 336 PHIVRYYDVMSDEEIERIKEIAKPKL--ARATVRDPKT-----GVLTVASYRVSKSSWLE 388
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQK----S 201
ED +A + ++ T + E + Y V +Y PH+D E K
Sbjct: 389 EDDDPVVARVNRRMQHITGLTVKTAELLQVANYGVGGQYEPHFDFSRNDERDTFKHLGTG 448
Query: 202 QRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNG 261
R+A+FL Y++DV+ GG T+FP +G + P++G + +Y+L +G
Sbjct: 449 NRVATFLNYMSDVEAGGATVFPD---------------LGAAIWPKKGTAVFWYNLLRSG 493
Query: 262 TIDPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
D + H +CPV+ G KWV+ KW + Q++
Sbjct: 494 EGDYRTRHAACPVLVGCKWVSNKWFHERGQEF 525
>P4H2_MOUSE (Q60716) Prolyl 4-hydroxylase alpha-2 subunit precursor
(EC 1.14.11.2) (4-PH alpha-2)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-2 subunit)
Length = 537
Score = 79.0 bits (193), Expect = 1e-14
Identities = 54/212 (25%), Positives = 98/212 (45%), Gaps = 30/212 (14%)
Query: 90 PRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS---- 145
P + + + + E+ + I +AK L + +R+ +T G+ T + VS S
Sbjct: 338 PHIVRYYDVMSDEEIERIKEIAKPKL--ARATVRDPKT-----GVLTVASYRVSKSSWLE 390
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQK----S 201
ED +A + ++ T + E + Y + +Y PH+D + K
Sbjct: 391 EDDDPVVARVNRRMQHITGLTVKTAELLQVANYGMGGQYEPHFDFSRSDDEDAFKRLGTG 450
Query: 202 QRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNG 261
R+A+FL Y++DV+ GG T+FP +G + P++G + +Y+L +G
Sbjct: 451 NRVATFLNYMSDVEAGGATVFPD---------------LGAAIWPKKGTAVFWYNLLRSG 495
Query: 262 TIDPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
D + H +CPV+ G KWV+ KW + Q++
Sbjct: 496 EGDYRTRHAACPVLVGCKWVSNKWFHERGQEF 527
>P4H2_CAEEL (Q20065) Prolyl 4-hydroxylase alpha-2 subunit precursor
(EC 1.14.11.2) (4-PH alpha-2)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-2 subunit)
Length = 539
Score = 79.0 bits (193), Expect = 1e-14
Identities = 56/221 (25%), Positives = 99/221 (44%), Gaps = 24/221 (10%)
Query: 78 ITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALA-LREGETEQNTKGIRT 136
+ I ++L + P A+ F N + + I +A LK + + + GE E T I
Sbjct: 313 LAPIKVEILRFDPLAVLFKNVIHDSEIEVIKELASPKLKRATVQNSKTGELEHATYRISK 372
Query: 137 SSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEY 196
S+ + D + + +I T + ++ E + Y + Y+PH+D E
Sbjct: 373 SAWL----KGDLDPVIDRVNRRIEDFTNLNQATSEELQVANYGLGGHYDPHFDFARKEEK 428
Query: 197 GPQKS----QRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGL 252
K+ R+A+ L Y++ + GG T+F + +G V P + D L
Sbjct: 429 NAFKTLNTGNRIATVLFYMSQPERGGATVF---------------NHLGTAVFPSKNDAL 473
Query: 253 LFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
+Y+L +G D + H +CPV+ G KWV+ KWI + Q++
Sbjct: 474 FWYNLRRDGEGDLRTRHAACPVLLGVKWVSNKWIHEKGQEF 514
>EGLX_MOUSE (Q8BG58) Putative HIF-prolyl hydroxylase PH-4 (EC
1.14.11.-) (Hypoxia-inducible factor prolyl
4-hydroxylase)
Length = 503
Score = 71.2 bits (173), Expect = 3e-12
Identities = 54/205 (26%), Positives = 83/205 (40%), Gaps = 47/205 (22%)
Query: 122 LREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRS---HGEAFNILRY 178
+R + E N +R S ++ E + I +++ R T + E ++RY
Sbjct: 262 MRSHKAESNEL-VRNSHHTWLHQGEGAHHVMRAIRQRVLRLTRLSPEIVEFSEPLQVVRY 320
Query: 179 EVDQRYNPHYDSFNPAEYGP--------------------QKSQRMASFLLYLTDVQEGG 218
Y+ H DS GP + S R + L YL +V GG
Sbjct: 321 GEGGHYHAHVDS------GPVYPETICSHTKLVANESVPFETSCRYMTVLFYLNNVTGGG 374
Query: 219 ETMFPFENGLNMDVSYRYEDCI------------GLRVRPRQGDGLLFYSLFPNGT---- 262
ET+FP + D +D + LRV+P+QG + +Y+ P+G
Sbjct: 375 ETVFPVADNRTYDEMSLIQDDVDLRDTRRHCDKGNLRVKPQQGTAVFWYNYLPDGQGWVG 434
Query: 263 -IDPTSLHGSCPVIKGEKWVATKWI 286
+D SLHG C V +G KW+A WI
Sbjct: 435 EVDDYSLHGGCLVTRGTKWIANNWI 459
>P4H1_CAEEL (Q10576) Prolyl 4-hydroxylase alpha-1 subunit precursor
(EC 1.14.11.2) (4-PH alpha-1)
(Procollagen-proline,2-oxoglutarate-4-dioxygenase
alpha-1 subunit)
Length = 559
Score = 69.3 bits (168), Expect = 1e-11
Identities = 50/218 (22%), Positives = 93/218 (41%), Gaps = 24/218 (11%)
Query: 81 IPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQN-TKGIRTSSG 139
I ++ + P A+ F + + ++ +I +AK L + + + T + T R S
Sbjct: 319 IKVEIKRFNPLAVLFKDVISDDEVAAIQELAKPKL--ARATVHDSVTGKLVTATYRISKS 376
Query: 140 VFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQ 199
++ E + + ++I T + E I Y + Y+PH+D E
Sbjct: 377 AWLKEWEGDV--VETVNKRIGYMTNLEMETAEELQIANYGIGGHYDPHFDHAKKEESKSF 434
Query: 200 KS----QRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFY 255
+S R+A+ L Y++ GG T+F + P + D L +Y
Sbjct: 435 ESLGTGNRIATVLFYMSQPSHGGGTVFTEAKST---------------ILPTKNDALFWY 479
Query: 256 SLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
+L+ G +P + H +CPV+ G KWV+ KWI + ++
Sbjct: 480 NLYKQGDGNPDTRHAACPVLVGIKWVSNKWIHEKGNEF 517
>EGLX_HUMAN (Q9NXG6) Putative HIF-prolyl hydroxylase PH-4 (EC
1.14.11.-) (Hypoxia-inducible factor prolyl
4-hydroxylase)
Length = 502
Score = 69.3 bits (168), Expect = 1e-11
Identities = 51/198 (25%), Positives = 82/198 (40%), Gaps = 46/198 (23%)
Query: 129 QNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSH---GEAFNILRYEVDQRYN 185
++++ +R S ++ E + I +++ R T + E ++RY Y+
Sbjct: 267 ESSELVRNSHHTWLYQGEGAHHIMRAIRQRVLRLTRLSPEIVELSEPLQVVRYGEGGHYH 326
Query: 186 PHYDSFNPAEYGP--------------------QKSQRMASFLLYLTDVQEGGETMFPFE 225
H DS GP + S R + L YL +V GGET+FP
Sbjct: 327 AHVDS------GPVYPETICSHTKLVANESVPFETSCRYMTVLFYLNNVTGGGETVFPVA 380
Query: 226 NGLNMDVSYRYEDCI------------GLRVRPRQGDGLLFYSLFPNGT-----IDPTSL 268
+ D +D + LRV+P+QG + +Y+ P+G +D SL
Sbjct: 381 DNRTYDEMSLIQDDVDLRDTRRHCDKGNLRVKPQQGTAVFWYNYLPDGQGWVGDVDDYSL 440
Query: 269 HGSCPVIKGEKWVATKWI 286
HG C V +G KW+A WI
Sbjct: 441 HGGCLVTRGTKWIANNWI 458
>CISY_SCHPO (Q10306) Probable citrate synthase, mitochondrial
precursor (EC 2.3.3.1)
Length = 473
Score = 32.7 bits (73), Expect = 1.1
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 2/73 (2%)
Query: 198 PQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDV--SYRYEDCIGLRVRPRQGDGLLFY 255
P MA F L +T ++ +E G+N Y YEDC+ L + G ++
Sbjct: 168 PPTLHPMAQFSLAVTALEHDSAFAKAYERGMNKHDYWKYEYEDCMDLIAKTVPIAGRIYR 227
Query: 256 SLFPNGTIDPTSL 268
+L+ +G + P +
Sbjct: 228 NLYRDGVVAPIQM 240
>PURA_BORPA (Q7W6Q7) Adenylosuccinate synthetase (EC 6.3.4.4)
(IMP--aspartate ligase) (AdSS) (AMPSase)
Length = 435
Score = 32.0 bits (71), Expect = 1.9
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 3/74 (4%)
Query: 98 FATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGT-LAIIE 156
F T+ C + A AG+ P AL G T+ T R SG F + D+ GT LA I
Sbjct: 242 FVTSSNCVAGAASAGAGVGPQALQYVLGITKAYT--TRVGSGPFPTELVDEIGTRLATIG 299
Query: 157 EKIARATMIPRSHG 170
++ T PR G
Sbjct: 300 KEFGSVTGRPRRCG 313
>PURA_BORBR (Q7WHP1) Adenylosuccinate synthetase (EC 6.3.4.4)
(IMP--aspartate ligase) (AdSS) (AMPSase)
Length = 435
Score = 32.0 bits (71), Expect = 1.9
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 3/74 (4%)
Query: 98 FATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGT-LAIIE 156
F T+ C + A AG+ P AL G T+ T R SG F + D+ GT LA I
Sbjct: 242 FVTSSNCVAGAASAGAGVGPQALQYVLGITKAYT--TRVGSGPFPTELVDEIGTRLATIG 299
Query: 157 EKIARATMIPRSHG 170
++ T PR G
Sbjct: 300 KEFGSVTGRPRRCG 313
>TERM_BPPRD (P09009) DNA terminal protein (Protein P8)
Length = 259
Score = 31.6 bits (70), Expect = 2.4
Identities = 24/82 (29%), Positives = 42/82 (50%), Gaps = 4/82 (4%)
Query: 105 KSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGTL---AIIEEKIAR 161
K + + KAGL P L +R+ + ++ KG+ S V++ K + A+IE AR
Sbjct: 19 KQVSNLKKAGLIPKTLDVRKVKPTKHYKGL-VSKYKDVATGGAKLAAIPNPAVIETLEAR 77
Query: 162 ATMIPRSHGEAFNILRYEVDQR 183
I + G+A+ R +++QR
Sbjct: 78 GESIIKKGGKAYLKARQQINQR 99
>HEM2_CHLPN (Q9Z7G1) Delta-aminolevulinic acid dehydratase (EC
4.2.1.24) (Porphobilinogen synthase) (ALAD) (ALADH)
Length = 332
Score = 31.6 bits (70), Expect = 2.4
Identities = 25/93 (26%), Positives = 38/93 (39%), Gaps = 3/93 (3%)
Query: 187 HYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYED--CIGLRV 244
H S + +Y + + L D +EG + + GL +DV YR C+ L
Sbjct: 215 HVTSGDKKQYQMNPKNVLEALLESSLDEEEGADILMVKPAGLYLDVIYRIRQNTCLPLAA 274
Query: 245 RPRQGDGLLFYSLFPNGTIDPTSL-HGSCPVIK 276
G+ + S F G +D +L H S IK
Sbjct: 275 YQVSGEYAMILSAFQQGWLDKETLFHESLIAIK 307
>RS5_HAEIN (P44374) 30S ribosomal protein S5
Length = 166
Score = 31.2 bits (69), Expect = 3.2
Identities = 19/73 (26%), Positives = 35/73 (47%), Gaps = 7/73 (9%)
Query: 120 LALREGETEQNTKGIRTSSGVFVSSSEDKTGTLA-----IIEEKIARATMIPRSHGEA-- 172
+AL EG + KG+ T S VF+ + + TG +A + E ++ +++G
Sbjct: 74 VALNEGTLQHPVKGVHTGSRVFMQPASEGTGIIAGGAMRAVLEVAGVRNVLSKAYGSTNP 133
Query: 173 FNILRYEVDQRYN 185
N++R +D N
Sbjct: 134 INVVRATIDALAN 146
>HUTH_BACSU (P10944) Histidine ammonia-lyase (EC 4.3.1.3)
(Histidase)
Length = 508
Score = 30.4 bits (67), Expect = 5.4
Identities = 21/67 (31%), Positives = 32/67 (47%), Gaps = 3/67 (4%)
Query: 98 FATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEE 157
F E+ ++ G+ KAG++P L +EG N T+ GV +K LA E
Sbjct: 162 FFEGERMPAMTGLKKAGIQPVTLTSKEGLALINGTQAMTAMGVVAYIEAEK---LAYQTE 218
Query: 158 KIARATM 164
+IA T+
Sbjct: 219 RIASLTI 225
>PURA_BORPE (Q7VWM1) Adenylosuccinate synthetase (EC 6.3.4.4)
(IMP--aspartate ligase) (AdSS) (AMPSase)
Length = 435
Score = 30.0 bits (66), Expect = 7.1
Identities = 25/74 (33%), Positives = 33/74 (43%), Gaps = 3/74 (4%)
Query: 98 FATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGT-LAIIE 156
F T+ C + A AG+ P AL G T+ T R SG F + D+ G LA I
Sbjct: 242 FVTSSNCVAGAASAGAGVGPQALQYVLGITKAYT--TRVGSGPFPTELVDEIGARLATIG 299
Query: 157 EKIARATMIPRSHG 170
++ T PR G
Sbjct: 300 KEFGSVTGRPRRCG 313
>PGCB_HUMAN (Q96GW7) Brevican core protein precursor (Brain enriched
hyaluronan binding protein) (BEHAB protein)
(UNQ2525/PRO6018)
Length = 911
Score = 30.0 bits (66), Expect = 7.1
Identities = 23/86 (26%), Positives = 41/86 (46%), Gaps = 3/86 (3%)
Query: 43 SQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAE 102
S+++G+ L + + EK E E D+++ + P ++ S P A P A+
Sbjct: 446 SEEEGKALEEEEKYEDEEEKEEEE--EEEEVEDEALWAWPSELSSPGPEAS-LPTEPAAQ 502
Query: 103 QCKSIVGVAKAGLKPSALALREGETE 128
+ A+A L+P A L +GE+E
Sbjct: 503 EKSLSQAPARAVLQPGASPLPDGESE 528
>AMHX_BACSU (P54983) Amidohydrolase amhX (EC 3.5.1.-) (Aminoacylase)
Length = 389
Score = 30.0 bits (66), Expect = 7.1
Identities = 21/63 (33%), Positives = 29/63 (45%), Gaps = 12/63 (19%)
Query: 215 QEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPV 274
++GG + E G+ D+ Y Y G+ VRP Q NG P+ LHGS
Sbjct: 126 EKGGGALKMIEEGVLDDIDYLY----GVHVRPIQET--------QNGRCAPSILHGSSQH 173
Query: 275 IKG 277
I+G
Sbjct: 174 IEG 176
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,558,936
Number of Sequences: 164201
Number of extensions: 1508623
Number of successful extensions: 3137
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 3099
Number of HSP's gapped (non-prelim): 25
length of query: 293
length of database: 59,974,054
effective HSP length: 109
effective length of query: 184
effective length of database: 42,076,145
effective search space: 7742010680
effective search space used: 7742010680
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0157b.6


