

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
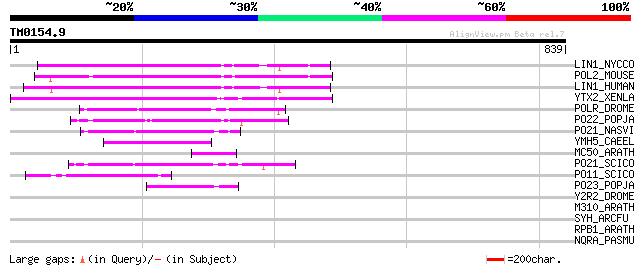
Query= TM0154.9
(839 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 155 5e-37
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 135 6e-31
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 135 6e-31
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 134 1e-30
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 73 4e-12
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 72 5e-12
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 67 3e-10
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 54 1e-06
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 53 4e-06
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 50 2e-05
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 49 6e-05
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 48 1e-04
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 44 0.001
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 41 0.012
SYH_ARCFU (O28631) Histidyl-tRNA synthetase (EC 6.1.1.21) (Histi... 36 0.48
RPB1_ARATH (P18616) DNA-directed RNA polymerase II largest subun... 32 9.1
NQRA_PASMU (Q9CLB1) Na(+)-translocating NADH-quinone reductase s... 32 9.1
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 155 bits (391), Expect = 5e-37
Identities = 116/461 (25%), Positives = 221/461 (47%), Gaps = 35/461 (7%)
Query: 42 EDPKVVKSKMKEFFEARFTNISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDC 101
E K++ K+ + ++ N+ LE +S+++ L R + EI + + +
Sbjct: 405 EIQKILNEYYKKLYSHKYENLKEIDQYLEACHLPRLSQKEVEMLNRPISSSEIASTIQNL 464
Query: 102 EGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKV-NNPM 160
KSPGPDG+ F ++F L ++ + ++ G+ P + ITLIPK +P
Sbjct: 465 PKKKSPGPDGFTSEFYQTFKEELVPILLNLFQNIEKEGILPNTFYEANITLIPKPGKDPT 524
Query: 161 GLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDE 220
+Y PISL+ KI++K+LT R+ + + II +Q F+ G Q ++ + VI
Sbjct: 525 RKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQH 584
Query: 221 A-KVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNG 279
K+K + ++ +D EK +D++ F+ L+++G ++ I+ + ++++NG
Sbjct: 585 INKLKNKDHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNG 644
Query: 280 SPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQF 339
F + G PL+P LF IV E L+ +R+ + + KG +G E +++SL F
Sbjct: 645 VKLKSFPLRSGTRQGCPLSPLLFNIVMEVLAIAIREEKAI---KGIHIGSEEIKLSL--F 699
Query: 340 ADDTLFIYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGVSVAEDVLLRYANLLHCK 399
ADD + E + + + +++ + +SG K+N KS V Y N +
Sbjct: 700 ADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKS----------VAFIYTNNNQAE 749
Query: 400 TT---EIPFV-------YLGIHVGANPRK--KTTWDPLLSKLRKRLSLWKRKTLSFGGKV 447
T IPF YLG+++ + + K ++ L ++ + ++ WK S+ G++
Sbjct: 750 KTVKDSIPFTVVPKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSWLGRI 809
Query: 448 CLIRSVLSSIPLFYLSFFKLP-KGVASLCNRIEK---QFLW 484
+++ +S +P +F +P K S +EK F+W
Sbjct: 810 NIVK--MSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIW 848
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 135 bits (339), Expect = 6e-31
Identities = 111/467 (23%), Positives = 218/467 (45%), Gaps = 27/467 (5%)
Query: 38 GGWVEDPKVVKSKMKEFFEARFT----NISGDGVLLEGTTFRSVSEEDNLALTRTFNLEE 93
G DP+ +++ ++ F++ ++ N+ L+ ++++ L + +E
Sbjct: 424 GDITTDPEEIQNTIRSFYKRLYSTKLENLDEMDKFLDRYQVPKLNQDQVDHLNSPISPKE 483
Query: 94 IRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLED-FH---TNGVWPKEGNSSF 149
I + KSPGPDG++ F ++F K+D++ +L FH G P +
Sbjct: 484 IEAVINSLPTKKSPGPDGFSAEFYQTF----KEDLIPILHKLFHKIEVEGTLPNSFYEAT 539
Query: 150 ITLIPKVN-NPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLL 208
ITLIPK +P + ++ PISL+ KI++K+L R+ + + II +Q F+ G Q
Sbjct: 540 ITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGW 599
Query: 209 DSVLVANEVIDEA-KVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGC 267
++ + VI K+K + ++ +D EK +D + F+ +L R G ++ IK
Sbjct: 600 FNIRKSINVIHYINKLKDKNHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMIKAI 659
Query: 268 IQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKV 327
++ VNG + ++ G PL+P+LF IV E L+ +RQ + + KG ++
Sbjct: 660 YSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQQKEI---KGIQI 716
Query: 328 GREGVEVSLLQFADDTLFIYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSK--LAGVSVA 385
G+E V++SLL ADD + ++ + +++ F + G K+N KS L +
Sbjct: 717 GKEEVKISLL--ADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLYTKNKQ 774
Query: 386 EDVLLRYANLLHCKTTEIPFVYLGIHVGANPRKKTTWDPLLSKLRKRLSLWKRKTLSFGG 445
+ +R T I ++ + + + L ++++ L WK S+ G
Sbjct: 775 AEKEIRETTPFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLPCSWIG 834
Query: 446 KVCLIRSVLSSIPLFYLSFFKLP-KGVASLCNRIEK---QFLWGGEE 488
++ +++ ++ +P F +P K N +E +F+W ++
Sbjct: 835 RINIVK--MAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVWNNKK 879
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 135 bits (339), Expect = 6e-31
Identities = 111/484 (22%), Positives = 221/484 (44%), Gaps = 36/484 (7%)
Query: 22 INWRRRTNAIRGLVVDGGWVE-DPKVVKSKMKEFFEARFTN----ISGDGVLLEGTTFRS 76
I +R N I + D G + DP +++ ++E+++ + N + L+ T
Sbjct: 380 IKKKREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLYANKLENLEEMDKFLDTYTLPR 439
Query: 77 VSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLEDFH 136
+++E+ +L R EI + KSPGP+G+ F + + L ++++ +
Sbjct: 440 LNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQSIE 499
Query: 137 TNGVWPKEGNSSFITLIPKVNNPMGLND-YIPISLIGCMYKIVSKVLTTRLSKVMGHIID 195
G+ P + I LIPK + + PISL+ KI++K+L ++ + + +I
Sbjct: 500 KEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIH 559
Query: 196 ENQSAFLEGRQLLDSVLVANEVIDEA-KVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRL 254
+Q F+ Q ++ + +I + K ++ +D EK +D + F+ L +L
Sbjct: 560 HDQVGFIPAMQGWFNIRKSINIIQHINRTKDTNHMIISIDAEKAFDKIQQPFMLKPLNKL 619
Query: 255 GFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMR 314
G ++ I+ + ++++NG + ++ G PL+P L IV E L+ +R
Sbjct: 620 GIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSPLLPNIVLEVLARAIR 679
Query: 315 QARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVLAMKSMLRCFELMSGLKVNF 374
Q + + KG ++G+E V++SL FADD + E + + ++ F +SG K+N
Sbjct: 680 QEKEI---KGIQLGKEEVKLSL--FADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINV 734
Query: 375 FKSKLAGVSVAEDVLLRYANLLHCKT---TEIPFV-------YLGIHVGANPRK--KTTW 422
KS+ Y N ++ +E+PF YLGI + + + K +
Sbjct: 735 QKSQ----------AFLYTNNRQTESQIMSELPFTIASKRIKYLGIQLTRDVKDLFKENY 784
Query: 423 DPLLSKLRKRLSLWKRKTLSFGGKVCLIRSVL--SSIPLFYLSFFKLPKGVASLCNRIEK 480
PLL+++++ + WK S+ G++ +++ + I F KLP + +
Sbjct: 785 KPLLNEIKEDTNKWKNIPCSWVGRINIVKMAILPKVIYRFNAIPIKLPMTFFTELEKTTL 844
Query: 481 QFLW 484
+F+W
Sbjct: 845 KFIW 848
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 134 bits (337), Expect = 1e-30
Identities = 125/496 (25%), Positives = 222/496 (44%), Gaps = 21/496 (4%)
Query: 2 KSRDRWVKDGDSNTKFFHNSINWRRRTNAIRGLVV-DGGWVEDPKVVKSKMKEFFEARFT 60
+SR + + D D ++FF+ + I L DG +EDP+ ++ + + F++ F+
Sbjct: 359 RSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNLFS 418
Query: 61 N--ISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIK 118
IS D VSE L L+E+ A+ +KSPG DG F +
Sbjct: 419 PDPISPDACEELWDGLPVVSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFFQ 478
Query: 119 SFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIV 178
FW L D RVL + G P + ++L+PK + + ++ P+SL+ YKIV
Sbjct: 479 FFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIV 538
Query: 179 SKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKRGFLVFKVDYEKV 238
+K ++ RL V+ +I +QS + GR + D+V + +++ A+ +D EK
Sbjct: 539 AKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFLSLDQEKA 598
Query: 239 YDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*MEKRLMLGDPLA 298
+D V+ +L L+ F +++G++K SA V +N S T + + G PL+
Sbjct: 599 FDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLS 658
Query: 299 PFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIYEPSTHNVLAMK 358
L+ + E ++R +RL G + + V L +ADD + + +++ ++
Sbjct: 659 GQLYSLAIEPFLCLLR--KRLT---GLVLKEPDMRVVLSAYADDVILV----AQDLVDLE 709
Query: 359 SMLRCFEL---MSGLKVNFFKSK-LAGVSVAEDVLLRYANLLHCKTTEIPFVYLGIHVGA 414
C E+ S ++N+ KS L S+ D L + ++ I YLG+++ A
Sbjct: 710 RAQECQEVYAAASSARINWSKSSGLLEGSLKVDFLPPAFRDISWESKIIK--YLGVYLSA 767
Query: 415 NPRK-KTTWDPLLSKLRKRLSLWK--RKTLSFGGKVCLIRSVLSSIPLFYLSFFKLPKGV 471
+ L + RL WK K LS G+ +I +++S + L +
Sbjct: 768 EEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRGRALVINQLVASQIWYRLICLSPTQEF 827
Query: 472 ASLCNRIEKQFLWGGE 487
+ R FLW G+
Sbjct: 828 IAKIQRRLLDFLWIGK 843
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 72.8 bits (177), Expect = 4e-12
Identities = 79/321 (24%), Positives = 135/321 (41%), Gaps = 23/321 (7%)
Query: 106 SPGPDGYNFHFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDY 165
SPGPDG KS V ++R++ G P + IPK D+
Sbjct: 359 SPGPDGIT---PKSAREVPSGIMLRIMNLILWCGNLPHSIRLARTVFIPKTVTAKRPQDF 415
Query: 166 IPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKK 225
PIS+ + + ++ +L TRL+ + D Q FL D+ + + V+ +
Sbjct: 416 RPISVPSVLVRQLNAILATRLNSSINW--DPRQRGFLPTDGCADNATIVDLVLRHSHKHF 473
Query: 226 RGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF 285
R + +D K +DS++ ++ LR G ++ +++ + S+ +G ++EF
Sbjct: 474 RSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQNTYEGGGTSLNGDGWSSEEF 533
Query: 286 *MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLF 345
+ + GDPL+P LF +V M R R L G KVG + FADD L
Sbjct: 534 VPARGVKQGDPLSPILFNLV------MDRLLRTLPSEIGAKVG--NAITNAAAFADD-LV 584
Query: 346 IYEPSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGV--SVAEDVLLRYANLLHCKTTEI 403
++ + + + F + GLK+N K G+ + + A + ++EI
Sbjct: 585 LFAETRMGLQVLLDKTLDFLSIVGLKLNADKCFTVGIKGQPKQKCTVLEAQSFYVGSSEI 644
Query: 404 P-------FVYLGIHVGANPR 417
P + YLGI+ A R
Sbjct: 645 PSLKRTDEWKYLGINFTATGR 665
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 72.4 bits (176), Expect = 5e-12
Identities = 84/339 (24%), Positives = 154/339 (44%), Gaps = 24/339 (7%)
Query: 92 EEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLEDFHT-NGVWPKEGNSSFI 150
EEI+ A+ + +PG DG I + + R H G P +
Sbjct: 9 EEIQCAIKGWK-PSAPGSDGLTVQAIT------RTRLPRNFVQLHLLRGHVPTPWTAMRT 61
Query: 151 TLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDS 210
TLIPK + +++ PI++ + +++ ++L RL + + A ++G L++S
Sbjct: 62 TLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVELHPAQKGYARIDGT-LVNS 120
Query: 211 VLVANEVIDEAKVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQS 270
+L+ + I + +++ + V +D K +D+V+ + + L+RLG +I G +
Sbjct: 121 LLL-DTYISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYITGSLSD 179
Query: 271 ASVSVLVN-GSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGR 329
++ ++ V GS T + + + + GDPL+PFLF V + L ++ + G +G
Sbjct: 180 STTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQSTPGI----GGTIGE 235
Query: 330 EGVEVSLLQFADDTLFIYE-----PSTHNVLAMKSMLRCFELMSGLKVNFFKSKLAGVSV 384
E ++ +L FADD L + + P+T +A LR L + V+ + GV +
Sbjct: 236 E--KIPVLAFADDLLLLEDNDVLLPTTLATVANFFRLRGMSLNAKKSVSISVAASGGVCI 293
Query: 385 AE-DVLLRYANLLHCKTTEI-PFVYLGIHVGANPRKKTT 421
LR N+L T + F YLG G + K T
Sbjct: 294 PRTKPFLRVDNVLLPVTDRMQTFRYLGHFFGLSGAAKPT 332
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 66.6 bits (161), Expect = 3e-10
Identities = 58/241 (24%), Positives = 107/241 (44%), Gaps = 14/241 (5%)
Query: 108 GPDGYNFHFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLIPKVNNPMGLNDYIP 167
GPDG + W+ + + I + +G P+ S LIPK M + P
Sbjct: 336 GPDGMT----TTAWNSIDECIKSLFNMIMYHGQCPRRYLDSRTVLIPKEPGTMDPACFRP 391
Query: 168 ISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKRG 227
+S+ + ++L R+ + ++D Q AF+ + ++ + + +I EA++K +G
Sbjct: 392 LSIASVALRHFHRILANRIGE--HGLLDTRQRAFIVADGVAENTSLLSAMIKEARMKIKG 449
Query: 228 FLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCIQSASVSVLVNGSPTDEF*M 287
+ +D +K +DSV + LRR + +I +++ + V +
Sbjct: 450 LYIAILDVKKAFDSVEHRSILDALRRKKLPLEMRNYIMWVYRNSKTRLEVVKTKGRWIRP 509
Query: 288 EKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVSLLQFADDTLFIY 347
+ + GDPL+P LF V + + RRL GF +G E ++ L FADD + +
Sbjct: 510 ARGVRQGDPLSPLLFNCVMDAV------LRRLPENTGFLMGAE--KIGALVFADDLVLLA 561
Query: 348 E 348
E
Sbjct: 562 E 562
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 54.3 bits (129), Expect = 1e-06
Identities = 36/164 (21%), Positives = 69/164 (41%), Gaps = 1/164 (0%)
Query: 142 PKEGNSSFITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAF 201
P + I IPK NP ++Y PISL +I+ +++ +R+ H++ +Q F
Sbjct: 662 PNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGF 721
Query: 202 LEGRQLLDSVLVANEVIDEAKVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWI 261
L R S++ + + ++ + D+ K +D V+ L L G
Sbjct: 722 LNFRSCPSSLVRSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTC 781
Query: 262 GWIKGCIQSASVSVLVNG-SPTDEF*MEKRLMLGDPLAPFLFLI 304
W K + + SV +N ++ + + + G P LF++
Sbjct: 782 SWFKEFLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFIL 825
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 52.8 bits (125), Expect = 4e-06
Identities = 25/68 (36%), Positives = 39/68 (56%)
Query: 276 LVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVGREGVEVS 335
++NG+P + L GDPL+P+LF++ E LSG+ R+A+ G +V ++
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRIN 72
Query: 336 LLQFADDT 343
L FADDT
Sbjct: 73 HLLFADDT 80
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 50.4 bits (119), Expect = 2e-05
Identities = 87/358 (24%), Positives = 147/358 (40%), Gaps = 37/358 (10%)
Query: 90 NLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLED-FHTNGVWPKEGNSS 148
+L EI++A +K GPDG W+ L D R+L + F G P S
Sbjct: 160 SLIEIKSA--RASNEKGAGPDGVT----PRSWNALDDRYKRLLYNIFVFYGRVPSPIKGS 213
Query: 149 FITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLL 208
PK+ + P+S+ + + +K+L R V + DE Q+A+L +
Sbjct: 214 RTVFTPKIEGGPDPGVFRPLSICSVILREFNKILARRF--VSCYTYDERQTAYLPIDGVC 271
Query: 209 DSVLVANEVIDEAKVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGCI 268
+V + +I EAK ++ + +D K ++SV + L + G + +I
Sbjct: 272 INVSMLTAIIAEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEAGCPPGVVDYIADMY 331
Query: 269 QSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVG 328
+ + G + + + GDPL+ LF + E + R LN F +
Sbjct: 332 NNVITEMQFEGK-CELASILAGVYQGDPLSGPLFTLAYE------KALRALNNEGRFDIA 384
Query: 329 REGVEVSLLQFADDTLFIYEPSTHNVLAMKSMLRCF-ELMS--GLKVNFFKSKLAGV--- 382
V V+ ++DD L + V+ ++ L F E ++ GL++N KSK +
Sbjct: 385 --DVRVNASAYSDDGLLL----AMTVIGLQHNLDKFGETLAKIGLRINSRKSKTVSLVPS 438
Query: 383 -------SVAEDVLLRYANLLHCKTTEIPFVYLG-IHVGANPR-KKTTWDPLLSKLRK 431
V+ LL A+ L T + YLG ++ + P K + D LSKL K
Sbjct: 439 GREKKMKIVSNRRLLLKASELKPLTISDLWKYLGVVYTTSGPEVAKVSMDDDLSKLTK 496
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 48.9 bits (115), Expect = 6e-05
Identities = 44/224 (19%), Positives = 102/224 (44%), Gaps = 16/224 (7%)
Query: 25 RRRTNAIRGLVVDGGWVEDPK-VVKSKMKEFFEARFTNISGDGVLLEGTTFRSVSEEDNL 83
RR+ + + VDG + + + V + M FF A + S E ++++
Sbjct: 350 RRKPVDVCSVKVDGVYTDTWEGSVNAMMNVFFPASIDDAS------EIDRLKAIARP--- 400
Query: 84 ALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLEDFHTNGVWPK 143
L ++E+ +V C+ KSPGPDG +++ W + + + + + +P+
Sbjct: 401 -LPPDLEMDEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQ 459
Query: 144 EGNSSFITLIPKVNNPMGLN--DYIPISLIGCMYKIVSKVLTTRL-SKVMGHIIDENQSA 200
+ + + ++ K+ + + + Y PI L+ + K++ ++ RL K+M + Q A
Sbjct: 460 KWKIASLVILLKLLDRIRSDPGSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFA 519
Query: 201 FLEGRQLLDSVLVANEVIDEAKVKKRGFLVFKVDYEKVYDSVNW 244
F G+ D+ ++ +++K + +D++ +D++ W
Sbjct: 520 FTYGKSTEDAWRCVQRHVECSEMKY--VIGLNIDFQGAFDNLGW 561
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 47.8 bits (112), Expect = 1e-04
Identities = 35/139 (25%), Positives = 64/139 (45%), Gaps = 6/139 (4%)
Query: 208 LDSVLVANEVIDEAKVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRLGFYAKWIGWIKGC 267
L ++++ I ++K + + V +D K +D+V+ + +R G +I
Sbjct: 3 LANIIMLEHYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMST 62
Query: 268 IQSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKV 327
I A +++V G T++ + + GDPL+P LF IV + L RLN +
Sbjct: 63 ITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDEL------VTRLNDEQPGAS 116
Query: 328 GREGVEVSLLQFADDTLFI 346
+++ L FADD L +
Sbjct: 117 MTPACKIASLAFADDLLLL 135
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 44.3 bits (103), Expect = 0.001
Identities = 79/355 (22%), Positives = 138/355 (38%), Gaps = 46/355 (12%)
Query: 93 EIRTAVWDCEGDKSPGPDGYNFHFIKSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITL 152
E+ T V + +SPG DG N K+ W + + + + G +P E +
Sbjct: 438 EVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVS 497
Query: 153 IPK--VNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDS 210
+ K + + Y I L+ K++ ++ R+ +V+ Q F +GR + D+
Sbjct: 498 LLKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPEGC-RWQFGFRQGRCVEDA 556
Query: 211 VLVANEVIDEAKVKKRGFLVFKVDYEKVYDSVNWNFLFYMLRRLGF--YAKWIGWIKGCI 268
+ + + L VD++ +D+V W+ L LG W + G
Sbjct: 557 WRHVKSSVGASAAQY--VLGTFVDFKGAFDNVEWSAALSRLADLGCREMGLWQSFFSG-- 612
Query: 269 QSASVSVLVNGSPTDEF*MEKRLMLGDPLAPFLFLIVAECLSGMMRQARRLNLYKGFKVG 328
+V+ + S T E + + G PF++ I+ + L +RL Y
Sbjct: 613 ---RRAVIRSSSGTVEVPVTRGCPQGSISGPFIWDILMDVL------LQRLQPY------ 657
Query: 329 REGVEVSLLQFADDTLFIYEPSTHNVLAMKSMLRCFELMS-----GLKVNFFKSKLAGVS 383
L +ADD L + E ++ VL K +LMS G +V S V
Sbjct: 658 -----CQLSAYADDLLLLVEGNSRAVLEEKGA----QLMSIVETWGAEVGDCLSTSKTVI 708
Query: 384 VAEDVLLRYANLLHCKTTEIPFV----YLGIHVGANPRKKTTWDPLLSKLRKRLS 434
+ LR A + +P+V YLGI V + T ++ LR+R++
Sbjct: 709 MLLKGALRRAPTVRFAGRNLPYVRSCRYLGITVSEGMKFLTH----IASLRQRMT 759
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 41.2 bits (95), Expect = 0.012
Identities = 16/48 (33%), Positives = 27/48 (55%)
Query: 456 SIPLFYLSFFKLPKGVASLCNRIEKQFLWGGEEGTRKIAWVKWSKVCR 503
++P++ +S F+L K + +F W E RKI+WV W K+C+
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCK 49
>SYH_ARCFU (O28631) Histidyl-tRNA synthetase (EC 6.1.1.21)
(Histidine--tRNA ligase) (HisRS)
Length = 409
Score = 35.8 bits (81), Expect = 0.48
Identities = 47/195 (24%), Positives = 82/195 (41%), Gaps = 19/195 (9%)
Query: 59 FTNISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCEGDKSPGPDGYNFHFIK 118
FT SG+G++ E F+ S D LAL +R V +C S P F++
Sbjct: 50 FTRKSGEGIIEEMYVFKDKSGRD-LALRPELTAPVMRMFVNEC----SVMPKPLRFYYFA 104
Query: 119 SFWHVLKDDIVRVLEDFHTN----GVWPKEGNSSFITLIPKVNNPMGLNDYIPISLIGCM 174
+ + + R E + G ++ I L K+ +G+N + I +G M
Sbjct: 105 NCFRYERPQKGRYREFWQFGVELIGSESYLADAEVIILADKILKDVGVNFSLEIGHVGIM 164
Query: 175 YKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLLDSVLVANEVIDEAKVKKRGFLVFKVD 234
++ + R SKVM +ID+ R+ L+S L V ++ + K L K D
Sbjct: 165 RHLLKPIGEDRASKVM-RLIDKGD------REGLESYLAEIRVNEDLRDKIFSLLELKGD 217
Query: 235 ---YEKVYDSVNWNF 246
E+ + ++++F
Sbjct: 218 ESVIEEAKEIIDYDF 232
>RPB1_ARATH (P18616) DNA-directed RNA polymerase II largest subunit
(EC 2.7.7.6)
Length = 1840
Score = 31.6 bits (70), Expect = 9.1
Identities = 54/218 (24%), Positives = 89/218 (40%), Gaps = 43/218 (19%)
Query: 51 MKEFFEARFTNISGDGVLLEGTTF-RSVSEEDNLALTRT-------FNLEEIRTAVW--D 100
M++ FE R + G++ +S++E +NL T N+ ++ V +
Sbjct: 730 MRDTFENRVNQVLNKARDDAGSSAQKSLAETNNLKAMVTAGSKGSFINISQMTACVGQQN 789
Query: 101 CEGDKSP-GPDGYNFHFIKSFWHVLKDDIVR----VLEDFHTNGVWPKE-------GNSS 148
EG + P G DG ++ H KDD +E+ + G+ P+E G
Sbjct: 790 VEGKRIPFGFDG------RTLPHFTKDDYGPESRGFVENSYLRGLTPQEFFFHAMGGREG 843
Query: 149 FITLIPKVNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQLL 208
I K + YI L+ M I+ K T + +G +I FL G +
Sbjct: 844 LIDTAVKTSE----TGYIQRRLVKAMEDIMVKYDGT-VRNSLGDVIQ-----FLYGEDGM 893
Query: 209 DSVLVANEVIDEAKVKKRGF-LVFKVDYEKVYDSVNWN 245
D+V + ++ +D K+KK F FK + D NWN
Sbjct: 894 DAVWIESQKLDSLKMKKSEFDRTFKYE----IDDENWN 927
>NQRA_PASMU (Q9CLB1) Na(+)-translocating NADH-quinone reductase
subunit A (EC 1.6.5.-) (Na(+)-translocating NQR subunit
A) (Na(+)-NQR subunit A) (NQR complex subunit A) (NQR-1
subunit A)
Length = 446
Score = 31.6 bits (70), Expect = 9.1
Identities = 39/172 (22%), Positives = 70/172 (40%), Gaps = 35/172 (20%)
Query: 43 DPKVVKSKMKEFFEARFTNISGDGVLLEGTTFRSVSEEDNLALTRTFNLEEIRTAVWDCE 102
DP+V+ + + FEA T +S L EG + + ++ NL+ V +
Sbjct: 162 DPQVIVQQSAQAFEAGLTVLSR---LHEGKVYLCKAANASIPSPSIANLD-----VKEFA 213
Query: 103 GDKSPGPDGYNFHFI------KSFWHVLKDDIVRVLEDFHTNGVWPKEGNSSFITLI-PK 155
G G G + HFI K W++ D++ V + F T + S ++L P+
Sbjct: 214 GPHPAGLSGTHIHFIDPVSATKFVWYINYQDVIAVGKLFTTGEL----DVSRVVSLAGPQ 269
Query: 156 VNNPMGLNDYIPISLIGCMYKIVSKVLTTRLSKVMGHIIDENQSAFLEGRQL 207
V NP ++V VL LS++ + + + ++ + G L
Sbjct: 270 VKNP----------------RLVRTVLGANLSQLTANEVKDGENRVISGSVL 305
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.337 0.148 0.491
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 94,270,620
Number of Sequences: 164201
Number of extensions: 3843608
Number of successful extensions: 10813
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 10777
Number of HSP's gapped (non-prelim): 22
length of query: 839
length of database: 59,974,054
effective HSP length: 119
effective length of query: 720
effective length of database: 40,434,135
effective search space: 29112577200
effective search space used: 29112577200
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0154.9


