

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
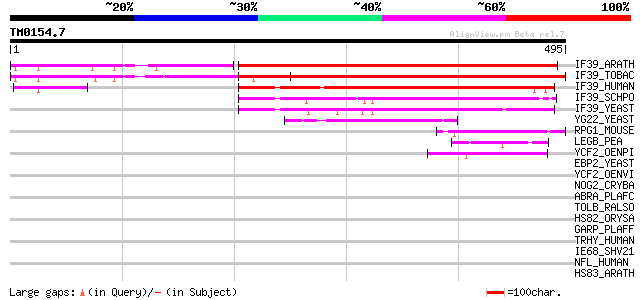
Query= TM0154.7
(495 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
IF39_ARATH (Q9C5Z1) Eukaryotic translation initiation factor 3 s... 460 e-129
IF39_TOBAC (P56821) Eukaryotic translation initiation factor 3 s... 431 e-120
IF39_HUMAN (P55884) Eukaryotic translation initiation factor 3 s... 205 2e-52
IF39_SCHPO (Q10425) Probable eukaryotic translation initiation f... 171 4e-42
IF39_YEAST (P06103) Eukaryotic translation initiation factor 3 9... 138 3e-32
YG22_YEAST (P53235) Hypothetical 71.3 kDa protein in SCM4-MUP1 i... 61 8e-09
RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase regulat... 46 3e-04
LEGB_PEA (P14594) Legumin B [Contains: Legumin B alpha chain (Le... 45 3e-04
YCF2_OENPI (P31568) Protein ycf2 (Fragment) 44 7e-04
EBP2_YEAST (P36049) rRNA processing protein EBP2 (EBNA1-binding ... 42 0.004
YCF2_OENVI (P31569) Protein ycf2 (Fragment) 41 0.006
NOG2_CRYBA (Q6TGJ8) Nucleolar GTP-binding protein 2 41 0.006
ABRA_PLAFC (P22620) 101 kDa malaria antigen (P101) (Acidic basic... 41 0.006
TOLB_RALSO (Q8Y1F5) TolB protein precursor 41 0.008
HS82_ORYSA (P33126) Heat shock protein 82 41 0.008
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 41 0.008
TRHY_HUMAN (Q07283) Trichohyalin 40 0.011
IE68_SHV21 (Q01042) Immediate-early protein 40 0.011
NFL_HUMAN (P07196) Neurofilament triplet L protein (68 kDa neuro... 40 0.014
HS83_ARATH (P51818) Heat shock protein 81-3 (HSP81-3) (HSP81.2) 40 0.014
>IF39_ARATH (Q9C5Z1) Eukaryotic translation initiation factor 3
subunit 9 (eIF-3 eta) (eIF3 p110) (eIF3b) (p82)
Length = 712
Score = 460 bits (1184), Expect = e-129
Identities = 218/285 (76%), Positives = 251/285 (87%), Gaps = 1/285 (0%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R+TKTKKSTY+GFELFRIKERDIPIEVLEL+NKNDKIIAFAWEPKGHRF VIHGD +
Sbjct: 425 VDRYTKTKKSTYSGFELFRIKERDIPIEVLELDNKNDKIIAFAWEPKGHRFAVIHGDQPR 484
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
PDVS YSM+TAQ G VSKL LK KQANALFWSP G++I++AGLKGFNGQLEF+NVDEL
Sbjct: 485 PDVSFYSMKTAQNTGRVSKLATLKAKQANALFWSPTGKYIILAGLKGFNGQLEFFNVDEL 544
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVTSVHDVENGFNIWSFSGKHLYRIMKDHFFQFSW 384
ETM TAEHFM TDIEWDPTGRYV T+VTSVH++ENGF IWSF+G LYRI+KDHFFQ +W
Sbjct: 545 ETMATAEHFMATDIEWDPTGRYVATAVTSVHEMENGFTIWSFNGIMLYRILKDHFFQLAW 604
Query: 385 RARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEW 444
R RPPSFLT EKEEEIA LKKYSKKYEAEDQD S+LLSEQ++EK + LKEEW+ WV +W
Sbjct: 605 RPRPPSFLTAEKEEEIAKTLKKYSKKYEAEDQDVSLLLSEQDREKRKALKEEWEKWVMQW 664
Query: 445 KWMHEEEKLQREQHRDGETSD-EEEEYEAKDIEVEEVIDVSEEIL 488
K +HEEEKL R+ RDGE SD EE+EYEAK++E E++IDV+EEI+
Sbjct: 665 KSLHEEEKLVRQNLRDGEVSDVEEDEYEAKEVEFEDLIDVTEEIV 709
Score = 94.7 bits (234), Expect = 5e-19
Identities = 74/222 (33%), Positives = 102/222 (45%), Gaps = 35/222 (15%)
Query: 1 MGY---KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPE 46
+GY ++NTPQEAQ AKE HGYKLD +FD M V +EW+PP+ PY
Sbjct: 105 LGYCFIEFNTPQEAQNAKEKSHGYKLDKSHIFAVNMFDDFDRLMNVKEEWEPPQARPYVP 164
Query: 47 KENQQHWLTDAKARDQFVIRTDSDTE---GRFPYKIKYLIHDLSFVRRS---GNSVADFL 100
EN Q WLTD KARDQ VIR+ DTE K +H + S + + +L
Sbjct: 165 GENLQKWLTDEKARDQLVIRSGPDTEVFWNDTRQKAPEPVHKRPYWTESYVQWSPLGTYL 224
Query: 101 AKNSSKFPNVVWLEEILPKFDLLVSSDAF--IVFAQVLVIMLLSMEEWQTIIEEFPKFDG 158
K VW +D F ++ Q ++ L+ + + + +
Sbjct: 225 V-TLHKQGAAVW-----------GGADTFTRLMRYQHSMVKLVDFSPGEKYLVTYHSQEP 272
Query: 159 VDPVSWIARAEKIFEVH-NLQEQDKSGCADEFAIGGTGGVTG 199
+P K+F+V +D G ADEF+IGG GGV G
Sbjct: 273 SNPRDASKVEIKVFDVRTGRMMRDFKGSADEFSIGGPGGVAG 314
>IF39_TOBAC (P56821) Eukaryotic translation initiation factor 3
subunit 9 (eIF-3 eta) (eIF3 p110) (eIF3b)
Length = 719
Score = 431 bits (1107), Expect = e-120
Identities = 205/293 (69%), Positives = 247/293 (83%), Gaps = 2/293 (0%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDS- 263
++R+TKTKKSTYTGFELFRIKERDIPIEVLEL+N NDKI AF W P+G +
Sbjct: 427 VDRYTKTKKSTYTGFELFRIKERDIPIEVLELDNTNDKIPAFGWGPEGSPICCYPSVTTP 486
Query: 264 KPDVSIYSMRT-AQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVD 322
+PD+ YS+ A G VSKLT LKGKQANAL+WSP GRF+++ GLKGFNGQLEF++VD
Sbjct: 487 RPDIRFYSVPVWALTPGLVSKLTTLKGKQANALYWSPGGRFLILTGLKGFNGQLEFFDVD 546
Query: 323 ELETMVTAEHFMTTDIEWDPTGRYVTTSVTSVHDVENGFNIWSFSGKHLYRIMKDHFFQF 382
ELETM +AEHFM TD+EWDPTGRYV TSVTSVH++ENGFNIWSF+GK LYRI+KDHFFQ+
Sbjct: 547 ELETMASAEHFMATDVEWDPTGRYVATSVTSVHEMENGFNIWSFNGKLLYRILKDHFFQY 606
Query: 383 SWRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVN 442
WR RPPSFL+ EKEEEIA NLK+YSKKYEAEDQD S+ LSEQ++EK + LKEEW+ W+N
Sbjct: 607 LWRPRPPSFLSKEKEEEIAKNLKRYSKKYEAEDQDVSLQLSEQDREKRKKLKEEWEAWIN 666
Query: 443 EWKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVIDVSEEILHFEYGQE 495
EWK +HEEEK++R++ RDGE SDEEEEYEAK++EVEE+I+V+EEI+ FE Q+
Sbjct: 667 EWKRLHEEEKMERQKLRDGEASDEEEEYEAKEVEVEEIINVTEEIIPFEESQQ 719
Score = 105 bits (262), Expect = 3e-22
Identities = 88/274 (32%), Positives = 133/274 (48%), Gaps = 35/274 (12%)
Query: 1 MGY---KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPE 46
+GY +YNTPQEA+L+KE HGYKLD + + F++VPDEW PPE +PY
Sbjct: 109 LGYCFIEYNTPQEAELSKEKTHGYKLDRSHIFAVNMFDDIERFLKVPDEWAPPEIKPYVP 168
Query: 47 KENQQHWLTDAKARDQFVIRTDSDTEGRF--PYKIK-YLIHDLSFVRRS---GNSVADFL 100
EN Q WLTD KARDQFVIR +DTE + ++K L++ SF S + + +L
Sbjct: 169 GENLQQWLTDEKARDQFVIRAGNDTEVLWNDARQLKPELVYKRSFWTESFVQWSPLGTYL 228
Query: 101 AKNSSKFPNVVWLEEILPKFDLLVSSDAFIVFAQVLVIMLLSMEEWQTIIEEFPKFDGVD 160
A + +W +S+ + +A V L+ + + + + +
Sbjct: 229 A-TVHRQGGAIW--------GGATTSNRLMRYAHPQV-KLIDFSLAERYLVTYSSHEPSN 278
Query: 161 PVSWIARAEKIFEVHNLQ-EQDKSGCADEFAIGGTGGVTGGRGLFLNRHTKTKKSTY--- 216
P A I +V + +D G ADEFA+GGTGGVTG R + K+ Y
Sbjct: 279 PRDSHAVVLNISDVRTGKVMRDFQGSADEFAVGGTGGVTGVSWPVF-RWSGGKQDKYFAR 337
Query: 217 TGFELFRIKERDIPIEVLELENKNDKIIAFAWEP 250
G + + E + + + K + ++ F+W P
Sbjct: 338 IGKNVISVYETETFSLIDKKSIKVENVMDFSWSP 371
>IF39_HUMAN (P55884) Eukaryotic translation initiation factor 3
subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b)
Length = 814
Score = 205 bits (521), Expect = 2e-52
Identities = 107/290 (36%), Positives = 177/290 (60%), Gaps = 14/290 (4%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 530 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPR 586
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 587 ISVSFYHVKNNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDC 643
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 644 TVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 703
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPP+ L+ E+ ++I +LKKYSK +E +D+ S++ E+ R + E++ +
Sbjct: 704 WRPRPPTLLSQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKM 763
Query: 444 WKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSEE 486
+ ++ E+K +R + R G +DE +++E + IE EE+I + +
Sbjct: 764 AQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVTEEIIPLGNQ 813
Score = 55.8 bits (133), Expect = 2e-07
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 236 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 295
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 296 WLEEAECRDQYSVIFES 312
>IF39_SCHPO (Q10425) Probable eukaryotic translation initiation
factor 3 90 kDa subunit (eIF3 p90)
Length = 725
Score = 171 bits (433), Expect = 4e-42
Identities = 115/306 (37%), Positives = 167/306 (53%), Gaps = 32/306 (10%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDS- 263
++RHTKTKKST++ E+FRI+E++IP+EV++L+ D ++ FAWEPK RF +I +D
Sbjct: 424 VDRHTKTKKSTFSNLEIFRIREKNIPVEVVDLK---DVVLNFAWEPKSDRFAIISANDQV 480
Query: 264 ------KPDVSIYSMRTAQKAGWVSK-LTALKGKQANALFWSPAGRFIVVAGLKGFNGQ- 315
K ++S Y + + + K N+LF +P GRF+V A L G + Q
Sbjct: 481 LNSTNVKTNLSFYGFEQKKNTPSTFRHIITFDKKTCNSLFMAPKGRFMVAATL-GSSTQY 539
Query: 316 -LEFYNVD------------ELETMVTAEHFMTTDIEWDPTGRYVTTSVTSV-HDVENGF 361
LEFY++D ++ + +AEHF T++EWDP+GRYVTTS T H +ENG+
Sbjct: 540 DLEFYDLDFDTEKKEPDALANVQQIGSAEHFGMTELEWDPSGRYVTTSSTIWRHKLENGY 599
Query: 362 NIWSFSGKHLYRIMKDHFFQFSWRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESML 421
+ F G L M F QF WR RPPS LT E ++I LK Y++ ++ ED E
Sbjct: 600 RLCDFRGTLLREEMIGEFKQFIWRPRPPSPLTKEDMKKIRKKLKDYNRLFDEEDIAEQSS 659
Query: 422 LSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVI 481
+ + + R L EW + +E EE+ Q ++EEE + VEEVI
Sbjct: 660 ANRELAARRRQLISEWQKYRDEVIARVAEERAITGQPAITVPAEEEEIIQE---TVEEVI 716
Query: 482 DVSEEI 487
SEEI
Sbjct: 717 --SEEI 720
Score = 36.2 bits (82), Expect = 0.20
Identities = 17/43 (39%), Positives = 26/43 (59%), Gaps = 1/43 (2%)
Query: 22 KLDEFDSFMRVPDEWDPPETEPYPEKENQQHWLTDAKARDQFV 64
KL++ + PDE+ E E + E+E+ + WLTD RDQF+
Sbjct: 126 KLNQLEKAFSTPDEFSFEERE-FKEREHLRSWLTDYYGRDQFI 167
>IF39_YEAST (P06103) Eukaryotic translation initiation factor 3 90
kDa subunit (eIF3 p90) (Cell cycle regulation and
translation initiation protein)
Length = 763
Score = 138 bits (348), Expect = 3e-32
Identities = 92/312 (29%), Positives = 158/312 (50%), Gaps = 35/312 (11%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
+ RHTK+ K+ ++ ++ R+ ERDIP+E +EL+ D + F WEP G+RFV I +
Sbjct: 454 VERHTKSGKTQFSNLQICRLTERDIPVEKVELK---DSVFEFGWEPHGNRFVTISVHEVA 510
Query: 265 P--------DVSIYSMRTAQKAGWVSKLTALKGKQ---ANALFWSPAGRFIVVAGLKGFN 313
+ Y+ T +K + + + +K AN + WSPAGRF+VV L G N
Sbjct: 511 DMNYAIPANTIRFYAPETKEKTDVIKRWSLVKEIPKTFANTVSWSPAGRFVVVGALVGPN 570
Query: 314 ---GQLEFYNVD---------------ELETMVTAEHFMTTDIEWDPTGRYVTTSVTSV- 354
L+FY++D L+ + + T+I WDP+GRYVT +S+
Sbjct: 571 MRRSDLQFYDMDYPGEKNINDNNDVSASLKDVAHPTYSAATNITWDPSGRYVTAWSSSLK 630
Query: 355 HDVENGFNIWSFSGKHLYRIMKDHFFQFSWRARPPSFLTPEKEEEIANNLKKYSKKYEAE 414
H VE+G+ I++ +G + + F F+WR RP S L+ + +++ NL+++S ++E +
Sbjct: 631 HKVEHGYKIFNIAGNLVKEDIIAGFKNFAWRPRPASILSNAERKKVRKNLREWSAQFEEQ 690
Query: 415 DQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHRDGETSDEEEEYEAKD 474
D E+ R L ++W + K E EK + D + D +++ +
Sbjct: 691 DAMEADTAMRDLILHQRELLKQWTEY--REKIGQEMEKSMNFKIFDVQPEDASDDFTTIE 748
Query: 475 IEVEEVIDVSEE 486
VEEV++ ++E
Sbjct: 749 EIVEEVLEETKE 760
Score = 34.3 bits (77), Expect = 0.76
Identities = 18/51 (35%), Positives = 25/51 (48%), Gaps = 6/51 (11%)
Query: 21 YKLDEFDSFMRVPDEWDPPETEPYPEKENQQHWLTDAKARDQFVIRTDSDT 71
Y D+FD+ R PD + + + WL D K RDQFV++ D T
Sbjct: 166 YNSDDFDTEFREPD------MPTFVPSSSLKSWLMDDKVRDQFVLQDDVKT 210
>YG22_YEAST (P53235) Hypothetical 71.3 kDa protein in SCM4-MUP1
intergenic region
Length = 642
Score = 60.8 bits (146), Expect = 8e-09
Identities = 40/156 (25%), Positives = 75/156 (47%), Gaps = 11/156 (7%)
Query: 246 FAWEPKGHRFVVIHGDDSKPDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIV 305
F W P +F VI G +S + +R + + +L + N + +SP+G +I+
Sbjct: 297 FTWSPTSRQFGVIAGY-MPATISFFDLRG-------NVVHSLPQQAKNTMLFSPSGHYIL 348
Query: 306 VAGLKGFNGQLEFYN-VDELETMVTAEHFMTTDIEWDPTGRYVTTSVTSVH-DVENGFNI 363
+AG G +E + +D+ + + + T+ +W P G ++ T+ TS V+NG I
Sbjct: 349 IAGFGNLQGSVEILDRLDKFKCVSKFDATNTSVCKWSPGGEFIMTATTSPRLRVDNGVKI 408
Query: 364 WSFSGKHLYRIMKDHFFQFSWRARPPSFLTPEKEEE 399
W SG ++ + WR+ P ++ T E ++E
Sbjct: 409 WHVSGSLVFVKEFKELLKVDWRS-PCNYKTLENKDE 443
>RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase
regulator-interacting protein 1 (RPGR-interacting
protein 1)
Length = 1331
Score = 45.8 bits (107), Expect = 3e-04
Identities = 32/119 (26%), Positives = 62/119 (51%), Gaps = 9/119 (7%)
Query: 381 QFSWRARPPSFLTPE----KEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEE 436
Q W++ +L PE E E +K + E E ++E + E+E+E+ +KEE
Sbjct: 886 QLDWKSH---YLAPEGFQMSEAEKPEGEEKEEEGGEEEVKEEEVEEEEEEEEEEEEVKEE 942
Query: 437 WDMWVNEWKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVIDVSEEILHFEYGQE 495
+ E + EE++ ++E+ + + +EEEE E ++ E EE D ++++L + +E
Sbjct: 943 KEEEEEEEREEEEEKEEEKEEEEEEDEKEEEEEEEEEEEEEEE--DENKDVLEASFTEE 999
Score = 40.8 bits (94), Expect = 0.008
Identities = 30/96 (31%), Positives = 46/96 (47%), Gaps = 3/96 (3%)
Query: 380 FQFSWRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDM 439
FQ S +P EKEEE K + E E+++E ++E+E+ + E +
Sbjct: 899 FQMSEAEKPEG---EEKEEEGGEEEVKEEEVEEEEEEEEEEEEVKEEKEEEEEEEREEEE 955
Query: 440 WVNEWKWMHEEEKLQREQHRDGETSDEEEEYEAKDI 475
E K EEE + E+ + E +EEEE E KD+
Sbjct: 956 EKEEEKEEEEEEDEKEEEEEEEEEEEEEEEDENKDV 991
Score = 39.3 bits (90), Expect = 0.024
Identities = 31/106 (29%), Positives = 52/106 (48%), Gaps = 9/106 (8%)
Query: 394 PEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKL 453
PE EE+ ++ K+ E E+++E E+E+E+ + KEE + E + EEEK
Sbjct: 907 PEGEEKEEEGGEEEVKEEEVEEEEEE----EEEEEEVKEEKEEEEEEEREEEEEKEEEKE 962
Query: 454 QREQH-----RDGETSDEEEEYEAKDIEVEEVIDVSEEILHFEYGQ 494
+ E+ + E +EEEE E ++ +V E E + F Q
Sbjct: 963 EEEEEDEKEEEEEEEEEEEEEEEDENKDVLEASFTEEWVPFFSQDQ 1008
>LEGB_PEA (P14594) Legumin B [Contains: Legumin B alpha chain
(Legumin B acidic chain); Legumin B beta chain (Legumin
B basic chain)] (Fragment)
Length = 338
Score = 45.4 bits (106), Expect = 3e-04
Identities = 32/91 (35%), Positives = 46/91 (50%), Gaps = 9/91 (9%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWD-----MWVNEWKWMHE 449
E+EEE +K+SKK E ED+DE +++ + KEE D + KW
Sbjct: 65 EEEEEQEQRHRKHSKK-EDEDEDEEEEEEREQRHRKHSEKEEEDEDEPRSYETRRKWKKH 123
Query: 450 EEKLQREQHRDGETSDEEEEYEAKDIEVEEV 480
+ +RE H GE EEEE E ++ E EE+
Sbjct: 124 TAEKKRESHGQGE---EEEELEKEEEEEEEI 151
>YCF2_OENPI (P31568) Protein ycf2 (Fragment)
Length = 721
Score = 44.3 bits (103), Expect = 7e-04
Identities = 31/114 (27%), Positives = 56/114 (48%), Gaps = 7/114 (6%)
Query: 373 RIMKDHFFQFSWRARPPSFLTPEKEEEIANNLKK-------YSKKYEAEDQDESMLLSEQ 425
RI F A P+ PE+E+E+ + + ++ EA+D+++ +L ++Q
Sbjct: 475 RIWHPWDFLLDCEAEIPAEEIPEEEDELPEDALETEVAVWGVEEEGEADDEEDVLLEAQQ 534
Query: 426 EQEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEE 479
E E EE D +E EE K + ++ + E +EEEE E ++ E++E
Sbjct: 535 EDELLEEEDEELDEEEDELDEEEEEPKEEEDELHEEEEEEEEEEEEEEEDELQE 588
>EBP2_YEAST (P36049) rRNA processing protein EBP2 (EBNA1-binding
protein homolog)
Length = 427
Score = 42.0 bits (97), Expect = 0.004
Identities = 25/88 (28%), Positives = 45/88 (50%), Gaps = 3/88 (3%)
Query: 401 ANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHRD 460
A+NLKK KK + D + + +E + + K+E E K M E++ + ++H
Sbjct: 45 ASNLKKLEKKEKKADVKKEVAADTEEYQSQALSKKEKRKLKKELKKMQEQDATEAQKHMS 104
Query: 461 G---ETSDEEEEYEAKDIEVEEVIDVSE 485
G E+ D+ EE E ++ E E +D+ +
Sbjct: 105 GDEDESGDDREEEEEEEEEEEGRLDLEK 132
>YCF2_OENVI (P31569) Protein ycf2 (Fragment)
Length = 630
Score = 41.2 bits (95), Expect = 0.006
Identities = 35/121 (28%), Positives = 58/121 (47%), Gaps = 16/121 (13%)
Query: 373 RIMKDHFFQFSWRARPPSFLTPEKEEEIANNLKK-------YSKKYEAEDQDESMLLSEQ 425
RI F A P+ PE+E+E+ + + ++ EA+D+++ +L ++Q
Sbjct: 386 RIWHPWDFLLDCEAEIPAEEIPEEEDELPEDALETEVAVWGVEEEGEADDEEDVLLEAQQ 445
Query: 426 EQEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVIDVSE 485
E E L EE D ++E EE++L E+ E DE E E ++ E EE D +
Sbjct: 446 EDE----LLEEEDEELDE-----EEDELDEEEEEPKEEEDELHEEEEEEEEEEEEEDELQ 496
Query: 486 E 486
E
Sbjct: 497 E 497
>NOG2_CRYBA (Q6TGJ8) Nucleolar GTP-binding protein 2
Length = 731
Score = 41.2 bits (95), Expect = 0.006
Identities = 22/88 (25%), Positives = 49/88 (55%)
Query: 400 IANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHR 459
I ++ ++Y ++ E DQD+ M+ E+E+E+ E+ + W+ + EE +
Sbjct: 575 IGDDARRYVEEEEVVDQDKEMVEKEEEEEEEEDEDEDEEEEELAWEDVFPEEADAVDGGE 634
Query: 460 DGETSDEEEEYEAKDIEVEEVIDVSEEI 487
+ E SD+EE + +D++ EE + ++++
Sbjct: 635 EVEESDKEETDDEEDVDEEEAVPSAKQL 662
>ABRA_PLAFC (P22620) 101 kDa malaria antigen (P101) (Acidic basic
repeat antigen)
Length = 743
Score = 41.2 bits (95), Expect = 0.006
Identities = 28/97 (28%), Positives = 52/97 (52%), Gaps = 6/97 (6%)
Query: 392 LTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEE 451
L E EEE+ ++ +K+ EAE + E+E+++ KE+ E K E+E
Sbjct: 643 LNQETEEEMEKQVEAITKQIEAEVDALAPKNKEEEEKEKEKEKEK------EEKEKEEKE 696
Query: 452 KLQREQHRDGETSDEEEEYEAKDIEVEEVIDVSEEIL 488
K ++E+ ++ + ++EE+ E K + EE + EEI+
Sbjct: 697 KEEKEKEKEEKEKEKEEKEEEKKEKEEEQEEEEEEIV 733
Score = 35.0 bits (79), Expect = 0.45
Identities = 24/88 (27%), Positives = 43/88 (48%), Gaps = 15/88 (17%)
Query: 392 LTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEE 451
L P+ +EE +K +K E E +++ E+E+EK E K +EE
Sbjct: 669 LAPKNKEEEEKEKEKEKEKEEKEKEEKEK--EEKEKEK-------------EEKEKEKEE 713
Query: 452 KLQREQHRDGETSDEEEEYEAKDIEVEE 479
K + ++ ++ E +EEEE +++ EE
Sbjct: 714 KEEEKKEKEEEQEEEEEEIVPENLTTEE 741
>TOLB_RALSO (Q8Y1F5) TolB protein precursor
Length = 432
Score = 40.8 bits (94), Expect = 0.008
Identities = 30/109 (27%), Positives = 52/109 (47%), Gaps = 6/109 (5%)
Query: 240 NDKIIAFAWEPKGHRFVVIHGDDSKPDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSP 299
N+ II+ AW P G + + + KP V ++ + + ++A ++ KG +A WSP
Sbjct: 197 NEPIISPAWSPDGTKVAYVSFESRKPVVYVHDLISGRRA----VISNQKGNN-SAPAWSP 251
Query: 300 AGRFIVVAGLKGFNGQLEFYNVDELE-TMVTAEHFMTTDIEWDPTGRYV 347
GR + VA + N Q+ N D +T + T+ + P GR +
Sbjct: 252 DGRRVAVALSRDGNTQIYQVNADGSGLRRLTRSSAIDTEPSFSPDGRSI 300
>HS82_ORYSA (P33126) Heat shock protein 82
Length = 699
Score = 40.8 bits (94), Expect = 0.008
Identities = 42/170 (24%), Positives = 74/170 (42%), Gaps = 19/170 (11%)
Query: 327 MVTAEHFMTTDIEWDPT--GRYVTTSVTSVHDVENGFNIWSF---------SGKHLYRIM 375
+VT +H W+ G + T TS + G I + + L ++
Sbjct: 137 VVTTKHNDDEQYVWESQAGGSFTVTRDTSGEQLGRGTKITLYLKDDQLEYLEERRLKDLI 196
Query: 376 KDHFFQFSWRARPPSFLTPEKE--EEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRML 433
K H S+ + T EKE ++ KK +++ + ED DE E++++K + +
Sbjct: 197 KKHSEFISYPISLWTEKTTEKEISDDEDEEEKKDAEEGKVEDVDEEKEEKEKKKKKIKEV 256
Query: 434 KEEWDMWVNEWK--WMHEEEKLQREQHR---DGETSDEEEEYEAKDIEVE 478
EW + VN+ K WM + E++ +E++ T+D EE K VE
Sbjct: 257 SHEWSL-VNKQKPIWMRKPEEITKEEYAAFYKSLTNDWEEHLAVKHFSVE 305
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 40.8 bits (94), Expect = 0.008
Identities = 27/85 (31%), Positives = 46/85 (53%)
Query: 402 NNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHRDG 461
N+ KK +K EAE Q + + E+++++ +E V E + EE++ + E+ +
Sbjct: 526 NHKKKMAKIEEAELQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEE 585
Query: 462 ETSDEEEEYEAKDIEVEEVIDVSEE 486
E +EEEE E ++ E EE D EE
Sbjct: 586 EEEEEEEEEEEEEEEEEEEEDEDEE 610
Score = 38.5 bits (88), Expect = 0.040
Identities = 25/92 (27%), Positives = 47/92 (50%), Gaps = 4/92 (4%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
E EEE+ + ++ ++ E E+++E E+E+E+ +EE D + E+E
Sbjct: 567 EDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEE----EEEEDEDEEDEDDAEEDEDDA 622
Query: 455 REQHRDGETSDEEEEYEAKDIEVEEVIDVSEE 486
E D E D+EE+ + +D + +E D +E
Sbjct: 623 EEDEDDAEEDDDEEDDDEEDDDEDEDEDEEDE 654
Score = 37.7 bits (86), Expect = 0.069
Identities = 21/92 (22%), Positives = 47/92 (50%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
+K++ + K + E +++ + + E+E E+ +EE + E + EEE+ +
Sbjct: 540 QKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEE 599
Query: 455 REQHRDGETSDEEEEYEAKDIEVEEVIDVSEE 486
E+ + E ++E++ E + + EE D +EE
Sbjct: 600 EEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEE 631
Score = 36.2 bits (82), Expect = 0.20
Identities = 22/85 (25%), Positives = 45/85 (52%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
E+EEE ++ ++ E E+++E E E+++ ++E D +E ++++
Sbjct: 578 EEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEED 637
Query: 455 REQHRDGETSDEEEEYEAKDIEVEE 479
++ D E DE+EE E ++ E EE
Sbjct: 638 DDEEDDDEDEDEDEEDEEEEEEEEE 662
Score = 35.8 bits (81), Expect = 0.26
Identities = 22/92 (23%), Positives = 47/92 (50%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
+KEE+ K+ ++ + +DE + ++E+E+ +EE + E + EEE+ +
Sbjct: 546 DKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 605
Query: 455 REQHRDGETSDEEEEYEAKDIEVEEVIDVSEE 486
E D + ++E+E+ +D + E D E+
Sbjct: 606 DEDEEDEDDAEEDEDDAEEDEDDAEEDDDEED 637
Score = 32.3 bits (72), Expect = 2.9
Identities = 23/93 (24%), Positives = 46/93 (48%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
E+EEE ++ ++ E E+++E E E E+ EE + E + EE+ +
Sbjct: 576 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDE 635
Query: 455 REQHRDGETSDEEEEYEAKDIEVEEVIDVSEEI 487
+ + + DE+E+ E ++ E EE + ++I
Sbjct: 636 EDDDEEDDDEDEDEDEEDEEEEEEEEEESEKKI 668
Score = 31.2 bits (69), Expect = 6.4
Identities = 20/92 (21%), Positives = 52/92 (55%), Gaps = 7/92 (7%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
+KE+E+ K KK + E++++ +QE+E+ + K+E + K M +++K++
Sbjct: 267 QKEKEMKEQEKIEKKKKKQEEKEKK----KQEKERKKQEKKERK---QKEKEMKKQKKIE 319
Query: 455 REQHRDGETSDEEEEYEAKDIEVEEVIDVSEE 486
+E+ + E ++++++ ++ E + D + E
Sbjct: 320 KERKKKEEKEKKKKKHDKENEETMQQPDQTSE 351
>TRHY_HUMAN (Q07283) Trichohyalin
Length = 1898
Score = 40.4 bits (93), Expect = 0.011
Identities = 29/100 (29%), Positives = 46/100 (46%), Gaps = 11/100 (11%)
Query: 385 RARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEW 444
R R L E+EE + LK+ +K +++ E L EQE+ + ++LK E +
Sbjct: 550 RLRSEQQLRREQEERLEQLLKREEEKRLEQERREQRLKREQEERRDQLLKREEERRQQRL 609
Query: 445 KWMHEE-----------EKLQREQHRDGETSDEEEEYEAK 473
K EE E+L++E+ RD EE E E +
Sbjct: 610 KREQEERLEQRLKREEVERLEQEERRDERLKREEPEEERR 649
Score = 39.7 bits (91), Expect = 0.018
Identities = 26/95 (27%), Positives = 50/95 (52%), Gaps = 7/95 (7%)
Query: 394 PEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWD-MWVNEWKWMHEEEK 452
PEK KKY ++ E + ++E +L E+E+ + ++EW+ + + + EEE+
Sbjct: 995 PEKRRRQERE-KKYREEEELQQEEEQLLREEREKRR----RQEWERQYRKKDELQQEEEQ 1049
Query: 453 LQREQHRDGETSDEEEEY-EAKDIEVEEVIDVSEE 486
L RE+ + E +Y E ++++ EE + EE
Sbjct: 1050 LLREEREKRRLQERERQYREEEELQQEEEQLLGEE 1084
Score = 37.7 bits (86), Expect = 0.069
Identities = 24/82 (29%), Positives = 41/82 (49%), Gaps = 5/82 (6%)
Query: 392 LTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQ-----EQEKCRMLKEEWDMWVNEWKW 446
L E+E L++ +K E Q+E LL E+ QE+ R +EE ++ E +
Sbjct: 1081 LGEERETRRRQELERQYRKEEELQQEEEQLLREEPEKRRRQERERQCREEEELQQEEEQL 1140
Query: 447 MHEEEKLQREQHRDGETSDEEE 468
+ EE + +R Q + + +EEE
Sbjct: 1141 LREEREKRRRQELERQYREEEE 1162
Score = 37.4 bits (85), Expect = 0.090
Identities = 21/68 (30%), Positives = 38/68 (55%), Gaps = 4/68 (5%)
Query: 405 KKYSKKYEAEDQDESMLLSEQE----QEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHRD 460
++Y KK E + ++E +L E+E QE+ R +EE ++ E + + EE + +R Q +
Sbjct: 1035 RQYRKKDELQQEEEQLLREEREKRRLQERERQYREEEELQQEEEQLLGEERETRRRQELE 1094
Query: 461 GETSDEEE 468
+ EEE
Sbjct: 1095 RQYRKEEE 1102
Score = 37.4 bits (85), Expect = 0.090
Identities = 29/109 (26%), Positives = 54/109 (48%), Gaps = 9/109 (8%)
Query: 396 KEEEIANNLKKYSKKYEAED---QDESMLLSEQ-----EQEKCRMLKEEWDMWVNEWKWM 447
+EE L++ ++Y E+ Q+E LL E+ QE R ++E ++ E + +
Sbjct: 1052 REEREKRRLQERERQYREEEELQQEEEQLLGEERETRRRQELERQYRKEEELQQEEEQLL 1111
Query: 448 HEEEKLQREQHRDGETSDEEE-EYEAKDIEVEEVIDVSEEILHFEYGQE 495
EE + +R Q R+ + +EEE + E + + EE + L +Y +E
Sbjct: 1112 REEPEKRRRQERERQCREEEELQQEEEQLLREEREKRRRQELERQYREE 1160
Score = 36.6 bits (83), Expect = 0.15
Identities = 24/87 (27%), Positives = 43/87 (48%), Gaps = 3/87 (3%)
Query: 396 KEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQR 455
+EE++ +K +K+ + + Q+E LL E+ +EK R + + E EE+
Sbjct: 1253 EEEQLEREEQKEAKRRDRKSQEEKQLLREEREEKRRRQETDRKFREEEQLLQEREEQPLL 1312
Query: 456 EQHRDGETSDEE---EEYEAKDIEVEE 479
Q RD + +EE +E K +E E+
Sbjct: 1313 RQERDRKFREEELLHQEQGRKFLEEEQ 1339
Score = 36.2 bits (82), Expect = 0.20
Identities = 19/79 (24%), Positives = 40/79 (50%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
E+ E+ ++ ++ + E++ E L EQE+ + + L+ E + E + E+E+ +
Sbjct: 313 ERREQQEERREQQERREQQEERREQQLRREQEERREQQLRREQEEERREQQLRREQEEER 372
Query: 455 REQHRDGETSDEEEEYEAK 473
REQ E +E E + +
Sbjct: 373 REQQLRREQEEERREQQLR 391
Score = 34.7 bits (78), Expect = 0.58
Identities = 28/100 (28%), Positives = 45/100 (45%), Gaps = 19/100 (19%)
Query: 394 PEKEEEIANNLKKYSKKYEAED---------------QDESMLLSEQEQEKCRMLKEEWD 438
PEKE + +N K Y K E E QD LL EQ++ R ++E
Sbjct: 1187 PEKENAVRDN-KVYCKGRENEQFRQLEDSQVRDRQSQQDLQHLLGEQQE---RDREQERR 1242
Query: 439 MWVNEWKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVE 478
W + EEE+L+RE+ ++ + D + + E + + E
Sbjct: 1243 RWQQANRHFPEEEQLEREEQKEAKRRDRKSQEEKQLLREE 1282
Score = 33.9 bits (76), Expect = 0.99
Identities = 25/96 (26%), Positives = 48/96 (49%), Gaps = 14/96 (14%)
Query: 392 LTPEKEEEIANNLKKYSKKYEAEDQ-----DESMLLSEQ------EQEKCRMLKEEWDM- 439
L PE+EE+ ++ +K+ E+Q +E L S++ E+E+ R +EE +
Sbjct: 1710 LRPEREEQQLRRQER-DRKFREEEQLRQGREEQQLRSQESDRKFREEEQLRQEREEQQLR 1768
Query: 440 -WVNEWKWMHEEEKLQREQHRDGETSDEEEEYEAKD 474
+ K+ EEE+LQ E+ + + +Y A++
Sbjct: 1769 PQQRDGKYRWEEEQLQLEEQEQRLRQERDRQYRAEE 1804
Score = 33.9 bits (76), Expect = 0.99
Identities = 23/101 (22%), Positives = 50/101 (48%), Gaps = 1/101 (0%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
++EEE L++ + E +++++ +EQ+ R +EE E + +E + Q
Sbjct: 272 QEEEEQLRKLERQELRRERQEEEQQQQRLRREQQLRRKQEEE-RREQQEERREQQERREQ 330
Query: 455 REQHRDGETSDEEEEYEAKDIEVEEVIDVSEEILHFEYGQE 495
+E+ R+ + E+EE + + E+ + E+ L E +E
Sbjct: 331 QEERREQQLRREQEERREQQLRREQEEERREQQLRREQEEE 371
Score = 32.3 bits (72), Expect = 2.9
Identities = 30/108 (27%), Positives = 50/108 (45%), Gaps = 13/108 (12%)
Query: 392 LTPEKEEEIANNLKKYSKKYEAEDQ--DESMLLSEQEQ------EKCRMLKEEWDMWVNE 443
L E+EEE+ ++ ++ E E Q +E L E+EQ EK R +E + +
Sbjct: 922 LLQEEEEELQREEREKRRRQEQERQYREEEQLQQEEEQLLREEREK-RRRQERERQYRKD 980
Query: 444 WKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVIDVSEEILHFE 491
K +EE+L E+ + E++Y E EE+ E++L E
Sbjct: 981 KKLQQKEEQLLGEEPEKRRRQEREKKYR----EEEELQQEEEQLLREE 1024
Score = 32.0 bits (71), Expect = 3.8
Identities = 18/88 (20%), Positives = 45/88 (50%), Gaps = 3/88 (3%)
Query: 395 EKEEEIANNLKKYSKKYEAED---QDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEE 451
+K E+ + ++ E D ++E EQE+ K ++ +++ + W + EEE
Sbjct: 455 QKHEQERREQRLKREQEERRDWLKREEETERHEQERRKQQLKRDQEEERRERWLKLEEEE 514
Query: 452 KLQREQHRDGETSDEEEEYEAKDIEVEE 479
+ ++++ R+ + E+EE + ++ +E
Sbjct: 515 RREQQERREQQLRREQEERREQRLKRQE 542
Score = 32.0 bits (71), Expect = 3.8
Identities = 23/84 (27%), Positives = 42/84 (49%), Gaps = 10/84 (11%)
Query: 397 EEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMW-VNEWKWMHEEEKLQR 455
EE+ ++ ++Y AE+Q + QEK R +EE ++W E K E E+ R
Sbjct: 1786 EEQEQRLRQERDRQYRAEEQFAT-------QEKSR--REEQELWQEEEQKRRQERERKLR 1836
Query: 456 EQHRDGETSDEEEEYEAKDIEVEE 479
E+H + +E+ + +I+ +E
Sbjct: 1837 EEHIRRQQKEEQRHRQVGEIQSQE 1860
Score = 31.6 bits (70), Expect = 4.9
Identities = 23/90 (25%), Positives = 40/90 (43%), Gaps = 2/90 (2%)
Query: 392 LTPEKEEEIANNLKK--YSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHE 449
L E+EE + LK+ + + E +DE + E E+E+ L + + + +
Sbjct: 609 LKREQEERLEQRLKREEVERLEQEERRDERLKREEPEEERRHELLKSEEQEERRHEQLRR 668
Query: 450 EEKLQREQHRDGETSDEEEEYEAKDIEVEE 479
E++ +REQ E +E E K EE
Sbjct: 669 EQQERREQRLKREEEEERLEQRLKREHEEE 698
Score = 31.2 bits (69), Expect = 6.4
Identities = 29/104 (27%), Positives = 50/104 (47%), Gaps = 19/104 (18%)
Query: 392 LTPEKEEEIANNLKKYSKKYEAE----DQDESMLLSEQ------EQEKCRMLKEEWDMWV 441
L E E+E ++Y K E E +++E L ++ E+E+ R +EE +
Sbjct: 1686 LRQETEQEQLRRQERYRKILEEEQLRPEREEQQLRRQERDRKFREEEQLRQGREEQQLRS 1745
Query: 442 NEW-KWMHEEEKLQRE--------QHRDGETSDEEEEYEAKDIE 476
E + EEE+L++E Q RDG+ EEE+ + ++ E
Sbjct: 1746 QESDRKFREEEQLRQEREEQQLRPQQRDGKYRWEEEQLQLEEQE 1789
Score = 30.8 bits (68), Expect = 8.4
Identities = 21/76 (27%), Positives = 40/76 (52%), Gaps = 2/76 (2%)
Query: 404 LKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHRDGET 463
L++ +K + Q+E L +E+EK R +E+ + E + EEE+L RE+
Sbjct: 912 LQEQLRKEQQLLQEEEEELQREEREK-RRRQEQERQYREEEQLQQEEEQLLREEREKRRR 970
Query: 464 SDEEEEYEAKDIEVEE 479
+ E +Y KD ++++
Sbjct: 971 QERERQYR-KDKKLQQ 985
Score = 30.8 bits (68), Expect = 8.4
Identities = 25/82 (30%), Positives = 41/82 (49%), Gaps = 7/82 (8%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
E EEE + L K E E++ L EQ++ + + LK E + E + E E+ +
Sbjct: 643 EPEEERRHELLKSE---EQEERRHEQLRREQQERREQRLKREEEEERLEQRLKREHEEER 699
Query: 455 REQHRDGETSDEEEEYEAKDIE 476
REQ E ++EE+E + I+
Sbjct: 700 REQ----ELAEEEQEQARERIK 717
>IE68_SHV21 (Q01042) Immediate-early protein
Length = 407
Score = 40.4 bits (93), Expect = 0.011
Identities = 28/95 (29%), Positives = 48/95 (50%), Gaps = 3/95 (3%)
Query: 395 EKEEEIANNLKKYSKKYEAED---QDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEE 451
E EEE A ++ +++ EAE+ ++E E E+E+ +EE E E E
Sbjct: 105 EAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAE 164
Query: 452 KLQREQHRDGETSDEEEEYEAKDIEVEEVIDVSEE 486
+ E+ + E ++EE E EA++ E E + +EE
Sbjct: 165 EEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEE 199
Score = 39.3 bits (90), Expect = 0.024
Identities = 28/101 (27%), Positives = 48/101 (46%), Gaps = 6/101 (5%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
E EEE A ++ +++ EAE++ +E+E E+ +EE + E + E E+ +
Sbjct: 145 EAEEEEAEEAEEEAEEEEAEEE------AEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAE 198
Query: 455 REQHRDGETSDEEEEYEAKDIEVEEVIDVSEEILHFEYGQE 495
E E ++E EE + E EE + EE E +E
Sbjct: 199 EEAEEAEEEAEEAEEEAEEAEEAEEAEEAEEEAEEAEEEEE 239
Score = 38.1 bits (87), Expect = 0.053
Identities = 26/94 (27%), Positives = 45/94 (47%), Gaps = 2/94 (2%)
Query: 395 EKEEEIANNLKKYSKKYEAED--QDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEK 452
E EEE A + ++ E E+ ++E+ E+E E+ EE + + E E+
Sbjct: 117 EAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEEAEEEAEEAEE 176
Query: 453 LQREQHRDGETSDEEEEYEAKDIEVEEVIDVSEE 486
+ E + E ++E EE E + E EE + +EE
Sbjct: 177 AEEEAEEEAEEAEEAEEAEEAEEEAEEAEEEAEE 210
Score = 37.7 bits (86), Expect = 0.069
Identities = 29/101 (28%), Positives = 47/101 (45%), Gaps = 1/101 (0%)
Query: 396 KEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQR 455
+EEE + + EAE+++ E E+E+ EE E + EEE +
Sbjct: 129 EEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEA 188
Query: 456 EQHRDGETSDEE-EEYEAKDIEVEEVIDVSEEILHFEYGQE 495
E+ + E ++EE EE E + E EE + +EE E +E
Sbjct: 189 EEAEEAEEAEEEAEEAEEEAEEAEEEAEEAEEAEEAEEAEE 229
Score = 37.0 bits (84), Expect = 0.12
Identities = 32/104 (30%), Positives = 53/104 (50%), Gaps = 5/104 (4%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLK-EEWDMWVNEWKWMHEEEKL 453
E EEE A +K +++ EAE+ +E E E+ + + EE + E + EEE
Sbjct: 95 EAEEEEAE--EKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAE 152
Query: 454 QREQHRDGETSDEEEEYEAKDIE--VEEVIDVSEEILHFEYGQE 495
+ E+ + E ++EE E EA++ E EE + +EE E +E
Sbjct: 153 EAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEE 196
Score = 36.2 bits (82), Expect = 0.20
Identities = 27/101 (26%), Positives = 44/101 (42%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
E EEE ++ ++ E E +E E E+E+ + E + + EEE +
Sbjct: 66 EVEEEGEERERRGEEEREGEGGEEGEGREEAEEEEAEEKEAEEEEAEEAEEEAEEEEAEE 125
Query: 455 REQHRDGETSDEEEEYEAKDIEVEEVIDVSEEILHFEYGQE 495
E + +E EE EA++ E EE + EE E +E
Sbjct: 126 AEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAEEEEAEEE 166
Score = 34.7 bits (78), Expect = 0.58
Identities = 26/101 (25%), Positives = 48/101 (46%), Gaps = 1/101 (0%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
E EE+ A + + EAE+++ +E+E+ + +EE E + EE+ +
Sbjct: 100 EAEEKEAEEEEAEEAEEEAEEEEAEEAEAEEEEAEEEEAEEEEAEEAEEEEAEEAEEEAE 159
Query: 455 REQHRDGETSDEEEEYEAKDIEVEEVIDVSEEILHFEYGQE 495
E+ + + EE EA++ E EE + +EE E +E
Sbjct: 160 EEEAEEEAEEEAEEAEEAEE-EAEEEAEEAEEAEEAEEAEE 199
Score = 34.3 bits (77), Expect = 0.76
Identities = 29/102 (28%), Positives = 53/102 (51%), Gaps = 8/102 (7%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQ 454
E EEE A ++ +++ E E+ +E+ +E+E+ +EE + E + EE + +
Sbjct: 132 EAEEEEAE--EEEAEEAEEEEAEEAEEEAEEEE-----AEEEAEEEAEEAEEAEEEAEEE 184
Query: 455 REQHRDGETSDE-EEEYEAKDIEVEEVIDVSEEILHFEYGQE 495
E+ + E ++E EEE E + E EE + +EE E +E
Sbjct: 185 AEEAEEAEEAEEAEEEAEEAEEEAEEAEEEAEEAEEAEEAEE 226
Score = 33.5 bits (75), Expect = 1.3
Identities = 27/100 (27%), Positives = 49/100 (49%), Gaps = 3/100 (3%)
Query: 395 EKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKE--EWDMWVNEWKWMHEEEK 452
E EEE + + EAE+ +E+ +E+E E+ +E E + E + EE +
Sbjct: 153 EAEEEAEEEEAEEEAEEEAEEAEEAEEEAEEEAEEAEEAEEAEEAEEEAEEAEEEAEEAE 212
Query: 453 LQREQHRDGETSDE-EEEYEAKDIEVEEVIDVSEEILHFE 491
+ E+ + E ++E EEE E + E EE + + H++
Sbjct: 213 EEAEEAEEAEEAEEAEEEAEEAEEEEEEAGPSTPRLPHYK 252
>NFL_HUMAN (P07196) Neurofilament triplet L protein (68 kDa
neurofilament protein) (Neurofilament light polypeptide)
(NF-L)
Length = 543
Score = 40.0 bits (92), Expect = 0.014
Identities = 35/137 (25%), Positives = 64/137 (46%), Gaps = 2/137 (1%)
Query: 344 GRYVTTSVTSVHDVENGFNIWS-FSGKHLYRIMKDHFFQFSWRARPPSFLTPEKEEEI-A 401
G S TSV + +G++ S G+ Y ++ + S R+ P + + +EE+
Sbjct: 395 GEETRLSFTSVGSITSGYSQSSQVFGRSAYGGLQTSSYLMSTRSFPSYYTSHVQEEQTEV 454
Query: 402 NNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKLQREQHRDG 461
+ SK EA+D+ S +E+E++ +EE E EE + E+ +G
Sbjct: 455 EETIEASKAEEAKDEPPSEGEAEEEEKDKEEAEEEEAAEEEEAAKEESEEAKEEEEGGEG 514
Query: 462 ETSDEEEEYEAKDIEVE 478
E +E +E E ++ +VE
Sbjct: 515 EEGEETKEAEEEEKKVE 531
>HS83_ARATH (P51818) Heat shock protein 81-3 (HSP81-3) (HSP81.2)
Length = 699
Score = 40.0 bits (92), Expect = 0.014
Identities = 27/90 (30%), Positives = 50/90 (55%), Gaps = 9/90 (10%)
Query: 397 EEEIANNLKKYSKKYE---AEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWK--WMHEEE 451
E+EI+++ ++ KK E E+ DE E++++K + + EWD+ VN+ K WM + E
Sbjct: 215 EKEISDDEEEEEKKDEEGKVEEVDEEKEKEEKKKKKIKEVSHEWDL-VNKQKPIWMRKPE 273
Query: 452 KLQREQHR---DGETSDEEEEYEAKDIEVE 478
++ +E++ ++D EE K VE
Sbjct: 274 EINKEEYAAFYKSLSNDWEEHLAVKHFSVE 303
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,742,720
Number of Sequences: 164201
Number of extensions: 2957912
Number of successful extensions: 14182
Number of sequences better than 10.0: 224
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 176
Number of HSP's that attempted gapping in prelim test: 12297
Number of HSP's gapped (non-prelim): 1063
length of query: 495
length of database: 59,974,054
effective HSP length: 114
effective length of query: 381
effective length of database: 41,255,140
effective search space: 15718208340
effective search space used: 15718208340
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0154.7


