

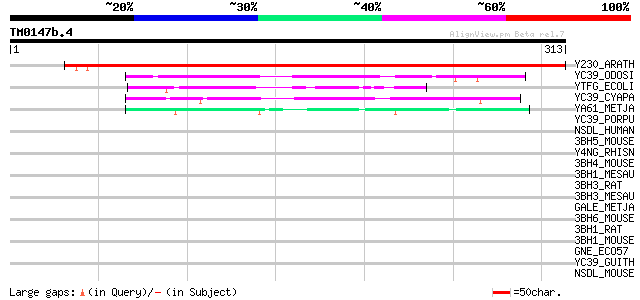
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0147b.4
(313 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
Y230_ARATH (O80934) Protein At2g37660, chloroplast precursor 432 e-121
YC39_ODOSI (P49534) Hypothetical 36.3 kDa protein ycf39 (ORF319) 57 5e-08
YTFG_ECOLI (P39315) Hypothetical protein ytfG 50 6e-06
YC39_CYAPA (P48279) Hypothetical 36.4 kDa protein ycf39 48 3e-05
YA61_METJA (Q58461) Hypothetical protein MJ1061 44 7e-04
YC39_PORPU (P51238) Hypothetical 35.7 kDa protein ycf39 (ORF319) 39 0.017
NSDL_HUMAN (Q15738) NAD(P)-dependent steroid dehydrogenase (EC 1... 37 0.049
3BH5_MOUSE (Q61694) 3 beta-hydroxysteroid dehydrogenase type V (... 37 0.063
Y4NG_RHISN (P55579) Hypothetical 42.8 kDa protein y4nG 36 0.11
3BH4_MOUSE (Q61767) 3 beta-hydroxysteroid dehydrogenase type IV ... 36 0.14
3BH1_MESAU (Q60555) 3 beta-hydroxysteroid dehydrogenase/delta 5-... 35 0.18
3BH3_RAT (P27364) 3 beta-hydroxysteroid dehydrogenase type III (... 35 0.31
3BH3_MESAU (O35296) 3 beta-hydroxysteroid dehydrogenase type III... 35 0.31
GALE_METJA (Q57664) Putative UDP-glucose 4-epimerase (EC 5.1.3.2... 34 0.41
3BH6_MOUSE (O35469) 3 beta-hydroxysteroid dehydrogenase/delta 5-... 34 0.41
3BH1_RAT (P22071) 3 beta-hydroxysteroid dehydrogenase/delta 5-->... 34 0.41
3BH1_MOUSE (P24815) 3 beta-hydroxysteroid dehydrogenase/delta 5-... 34 0.41
GNE_ECO57 (Q8X7P7) UDP-N-acetylglucosamine 4-epimerase (EC 5.1.3... 34 0.54
YC39_GUITH (O78472) Hypothetical 35.9 kDa protein ycf39 33 0.70
NSDL_MOUSE (Q9R1J0) NAD(P)-dependent steroid dehydrogenase (EC 1... 33 0.70
>Y230_ARATH (O80934) Protein At2g37660, chloroplast precursor
Length = 325
Score = 432 bits (1112), Expect = e-121
Identities = 216/289 (74%), Positives = 253/289 (86%), Gaps = 7/289 (2%)
Query: 32 TSLQF----SHSSSL---SLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKL 84
+SLQF S S+S+ S T K R +V A A + TVLVTGAGGRTG+IVYKKL
Sbjct: 37 SSLQFRSLVSDSTSICGPSKFTGKNRRVSVTVSAAATTEPLTVLVTGAGGRTGQIVYKKL 96
Query: 85 QERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMK 144
+ERS Q++ARGLVRT+ESK+ I D+V++GDIRDT SIAPA++GIDAL+ILTSAVP MK
Sbjct: 97 KERSEQFVARGLVRTKESKEKINGEDEVFIGDIRDTASIAPAVEGIDALVILTSAVPQMK 156
Query: 145 PGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLN 204
PGF+P+KG RPEF+F+DGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGT++N+PLN
Sbjct: 157 PGFDPSKGGRPEFFFDDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTNINHPLN 216
Query: 205 SLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIA 264
S+GN NILVWKRKAEQYLADSGIPYTIIRAGGLQDK+GG+REL++GKDDE+L+TETRTIA
Sbjct: 217 SIGNANILVWKRKAEQYLADSGIPYTIIRAGGLQDKDGGIRELLVGKDDELLETETRTIA 276
Query: 265 RPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFSQITTRF 313
R DVAEVC+QAL EEA+FKA DLASKPEG GTPT+DFKALF+Q+TT+F
Sbjct: 277 RADVAEVCVQALQLEEAKFKALDLASKPEGTGTPTKDFKALFTQVTTKF 325
>YC39_ODOSI (P49534) Hypothetical 36.3 kDa protein ycf39 (ORF319)
Length = 319
Score = 57.4 bits (137), Expect = 5e-08
Identities = 64/234 (27%), Positives = 99/234 (41%), Gaps = 38/234 (16%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGA-SDDVYVGDIRDTGSIA 124
++L+ G G G+ V LQ + Y R LVR + ++ GD+ +I
Sbjct: 2 SLLIIGGTGTLGRQVV--LQALTKGYQVRCLVRNFRKANFLKEWGAELIYGDLSRPETIP 59
Query: 125 PAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAG 184
P +QGI A+I +++ P D +QVDW G+ I+AA+AA
Sbjct: 60 PCLQGITAVIDTSTSRP------------------SDLDTLKQVDWDGKCALIEAAQAAN 101
Query: 185 VKQIVLVGSMGGTD-LNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGG 243
VK V S LN PL + G E L S IPYT+ R G +G
Sbjct: 102 VKHFVFCSSQNVEQFLNIPLMEMKFG--------IETKLQQSNIPYTVFRLAGFY--QGL 151
Query: 244 VRELIIG--KDDEILKTETRT----IARPDVAEVCIQALNFEEAQFKAFDLASK 291
+ + I ++ IL T T + D+A+ C+++L E + + F L +
Sbjct: 152 IEQYAIPVLENLPILVTNENTCVSYMDTQDIAKFCLRSLQLPETKNRTFVLGGQ 205
>YTFG_ECOLI (P39315) Hypothetical protein ytfG
Length = 286
Score = 50.4 bits (119), Expect = 6e-06
Identities = 51/172 (29%), Positives = 80/172 (45%), Gaps = 38/172 (22%)
Query: 67 VLVTGAGGRTGKIVYKKLQER--SSQYIARGLVRTEESKQTIGASD-DVYVGDIRDTGSI 123
+ +TGA G+ G V + L + +SQ +A +VR Q + A V D D ++
Sbjct: 2 IAITGATGQLGHYVIESLMKTVPASQIVA--IVRNPAKAQALAAQGITVRQADYGDEAAL 59
Query: 124 APAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAA 183
A+QG++ L++++S+ E G Q +N I+AAKAA
Sbjct: 60 TSALQGVEKLLLISSS--------------------EVGQRAPQ-----HRNVINAAKAA 94
Query: 184 GVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAG 235
GVK I + +PL L + +I + E+ LADSGI YT++R G
Sbjct: 95 GVKFIAYTSLLHAD--TSPLG-LADEHI-----ETEKMLADSGIVYTLLRNG 138
>YC39_CYAPA (P48279) Hypothetical 36.4 kDa protein ycf39
Length = 321
Score = 48.1 bits (113), Expect = 3e-05
Identities = 59/231 (25%), Positives = 97/231 (41%), Gaps = 38/231 (16%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTI---GASDDVYVGDIRDTGS 122
++LV GA G G+ + + + Y R LVR + GA + GD+ S
Sbjct: 2 SILVIGATGTLGRQIVRSALDEG--YQVRCLVRNLRKAAFLKEWGAK--LIWGDLSQPES 57
Query: 123 IAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKA 182
+ PA+ GI +I +++ P D A QVD G+K IDAAKA
Sbjct: 58 LLPALTGIRVIIDTSTSRPT------------------DPAGVYQVDLKGKKALIDAAKA 99
Query: 183 AGVKQIVLVGSMGGTDLNN-PLNSLGNGNILVWKRKAEQYLADSGIPYTIIR-AGGLQDK 240
+++ + + + PL + K E+ L +SG+ YTI + G Q
Sbjct: 100 MKIEKFIFFSILNSEKYSQVPLMRI--------KTVTEELLKESGLNYTIFKLCGFFQGL 151
Query: 241 EGGVRELIIGKDDEILKTETRTIA---RPDVAEVCIQALNFEEAQFKAFDL 288
G I+ + + TE+ +IA D+A +++L +E + F L
Sbjct: 152 IGQYAVPILDQQTVWITTESTSIAYMDTIDIARFTLRSLVLKETNNRVFPL 202
>YA61_METJA (Q58461) Hypothetical protein MJ1061
Length = 333
Score = 43.5 bits (101), Expect = 7e-04
Identities = 51/236 (21%), Positives = 95/236 (39%), Gaps = 29/236 (12%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQYI----ARGLVRTEESKQTIGASDDVYVGDIRDTG 121
T+LVTG G GK + K L + + + I E + ++GD+RD
Sbjct: 13 TILVTGGTGSIGKEIVKTLLKFNPKTIRVLDINETALFELEHELNSEKIRCFIGDVRDKD 72
Query: 122 SIAPAIQGIDALIILTSA--VPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDA 179
+ AI+ +D + + VPL + +NP + + + IG +N I+
Sbjct: 73 RLKRAIEEVDVVFHAAALKHVPLCE--YNPFEAVK-------------TNVIGTQNLIEV 117
Query: 180 AKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKR--KAEQYLADSGIPYTIIRAGGL 237
A V++ + + + NP+N +G +L + A Y ++++R G +
Sbjct: 118 AMDEEVEKFITISTDKAV---NPVNVMGATKLLAERLTISANLYKGKRKTAFSVVRFGNV 174
Query: 238 QDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPE 293
+ G + L+ ++I K T+ PD+ + + KA LA E
Sbjct: 175 LNSRGSILPLL---KEQIKKGGPVTLTHPDMTRFIMSINEAVKLVLKACYLAKGGE 227
>YC39_PORPU (P51238) Hypothetical 35.7 kDa protein ycf39 (ORF319)
Length = 319
Score = 38.9 bits (89), Expect = 0.017
Identities = 42/173 (24%), Positives = 71/173 (40%), Gaps = 30/173 (17%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGA-SDDVYVGDIRDTGSIA 124
T+LV GA G G+ + ++ + Y + +VR + ++ GD++ SI
Sbjct: 2 TLLVIGATGTLGRQIVRRALDEG--YNVKCMVRNLRKSAFLKEWGAELVYGDLKLPESIL 59
Query: 125 PAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAG 184
+ G+ A+I +++ P D EQ+D G+ I+AAKAA
Sbjct: 60 QSFCGVTAVIDASTSRP------------------SDPYNTEQIDLDGKIALIEAAKAAK 101
Query: 185 VKQIVLVGSMGGTDLNN-PLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGG 236
V++ + + PL +L K + YL S I YT+ GG
Sbjct: 102 VQRFIFFSILNADQYPKVPLMNL--------KSQVVNYLQKSSISYTVFSLGG 146
>NSDL_HUMAN (Q15738) NAD(P)-dependent steroid dehydrogenase (EC
1.1.1.-) (H105e3 protein)
Length = 373
Score = 37.4 bits (85), Expect = 0.049
Identities = 25/98 (25%), Positives = 45/98 (45%), Gaps = 20/98 (20%)
Query: 113 YVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIG 172
++GD+ + PA++G++ + S P + + FY +V++IG
Sbjct: 81 FLGDLCSRQDLYPALKGVNTVFHCASPPP--------SSNNKELFY--------RVNYIG 124
Query: 173 QKNQIDAAKAAGVKQIVLVGSMG----GTDLNNPLNSL 206
KN I+ K AGV++++L S G D+ N L
Sbjct: 125 TKNVIETCKEAGVQKLILTSSASVIFEGVDIKNGTEDL 162
>3BH5_MOUSE (Q61694) 3 beta-hydroxysteroid dehydrogenase type V
(3Beta-HSD V) (3-beta-hydroxy-delta(5)-steroid
dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene
steroid dehydrogenase) (Progesterone reductase)
Length = 372
Score = 37.0 bits (84), Expect = 0.063
Identities = 28/80 (35%), Positives = 37/80 (46%), Gaps = 7/80 (8%)
Query: 68 LVTGAGGRTGKIVYKKLQERSSQYIARGLVRT-------EESKQTIGASDDVYVGDIRDT 120
LVTGAGG G+ + + L + R L RT E SK A V GDI D
Sbjct: 6 LVTGAGGFLGQRIVRMLVQEEELQEIRALFRTFGRKHEEELSKLQTKAKVRVLKGDILDA 65
Query: 121 GSIAPAIQGIDALIILTSAV 140
+ A QG+ A+I +A+
Sbjct: 66 QCLKRACQGMSAVIHTAAAI 85
>Y4NG_RHISN (P55579) Hypothetical 42.8 kDa protein y4nG
Length = 396
Score = 36.2 bits (82), Expect = 0.11
Identities = 42/172 (24%), Positives = 74/172 (42%), Gaps = 26/172 (15%)
Query: 38 HSSSLSLATHKRVR-TRTSVVAMA---DSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIA 93
H +L + T + VR ++TS++A+ T LVTG G G+++ K+L + +
Sbjct: 19 HGKTLQIPTLRWVRQSKTSIMALVICLGRIVKTALVTGGSGYFGELLSKQLLRQGTYVRV 78
Query: 94 RGLVRTEESKQTIGASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGE 153
L S + + G I D ++ A+ GID + + VPL K E
Sbjct: 79 FDLNPPGFSHPNL----EFLKGTILDRNAVRQALSGIDKVFHNVAQVPLAK--------E 126
Query: 154 RPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMG--GTDLNNPL 203
+ F+ V+ G + +D + A G+++ V S G +NP+
Sbjct: 127 KDLFW--------SVNCGGTQIIVDESVATGIEKFVYTSSSAVFGAPKSNPV 170
>3BH4_MOUSE (Q61767) 3 beta-hydroxysteroid dehydrogenase type IV
(3Beta-HSD IV) (3-beta-hydroxy-delta(5)-steroid
dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene
steroid dehydrogenase) (Progesterone reductase)
Length = 372
Score = 35.8 bits (81), Expect = 0.14
Identities = 27/80 (33%), Positives = 36/80 (44%), Gaps = 7/80 (8%)
Query: 68 LVTGAGGRTGKIVYKKLQERSSQYIARGLVRT-------EESKQTIGASDDVYVGDIRDT 120
LVTGAGG G+ + + L + R L RT E SK V GDI D
Sbjct: 6 LVTGAGGFLGQRIVRMLVQEEELQEIRALFRTFGRKQEEELSKLQTKTKVTVLKGDILDA 65
Query: 121 GSIAPAIQGIDALIILTSAV 140
+ A QG+ A+I +A+
Sbjct: 66 QCLKRACQGMSAVIHTAAAI 85
>3BH1_MESAU (Q60555) 3 beta-hydroxysteroid dehydrogenase/delta
5-->4-isomerase type I (3Beta-HSD I) [Includes:
3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC
1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase)
(Progesterone reductase); Steroid delt
Length = 372
Score = 35.4 bits (80), Expect = 0.18
Identities = 49/196 (25%), Positives = 74/196 (37%), Gaps = 44/196 (22%)
Query: 68 LVTGAGGRTGKIVY------KKLQE-RSSQYIARGLVRTEESKQTIGASDDVYVGDIRDT 120
LVTGAGG G+ + K+LQE R+ + R R E K V GDI D
Sbjct: 6 LVTGAGGFLGQRIIRMLVQEKELQEVRALDKVFRPETREEFCKLQTKTKVTVLEGDILDA 65
Query: 121 GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQ----VDWIGQKNQ 176
+ A QGI +I +A+ + GA P Q ++ G N
Sbjct: 66 QCLRRACQGISVVIHTAAAIDVF------------------GAIPRQTIIDINLKGTLNL 107
Query: 177 IDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPY------- 229
++A A V + S+ N+ + NG+ + +Q+ + PY
Sbjct: 108 LEACVQASVPAFIYTSSIDVAGPNSYKEIVLNGH------EEQQHESTWSDPYPYSKKMA 161
Query: 230 --TIIRAGGLQDKEGG 243
++ A G K GG
Sbjct: 162 EKAVLAANGSSLKNGG 177
>3BH3_RAT (P27364) 3 beta-hydroxysteroid dehydrogenase type III
(3Beta-HSD III) (3-beta-hydroxy-delta(5)-steroid
dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene
steroid dehydrogenase) (Progesterone reductase)
Length = 372
Score = 34.7 bits (78), Expect = 0.31
Identities = 35/90 (38%), Positives = 44/90 (48%), Gaps = 11/90 (12%)
Query: 68 LVTGAGGRTG-KIVYKKLQERSSQYIARGLVRT-------EESKQTIGASDDVYVGDIRD 119
LVTGAGG G +IV +QE+ Q + R L RT E SK A V GDI D
Sbjct: 6 LVTGAGGFLGQRIVQMLVQEKELQEV-RVLYRTFSPKHKEELSKLQTKAKVTVLRGDIVD 64
Query: 120 TGSIAPAIQGIDALIILTSAVPLMKPGFNP 149
+ A QG+ +I+ +A L GF P
Sbjct: 65 AQFLRRACQGMS--VIIHTAAALDIAGFLP 92
>3BH3_MESAU (O35296) 3 beta-hydroxysteroid dehydrogenase type III
(3Beta-HSD III) (3-beta-hydroxy-delta(5)-steroid
dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene
steroid dehydrogenase) (Progesterone reductase)
Length = 372
Score = 34.7 bits (78), Expect = 0.31
Identities = 47/196 (23%), Positives = 73/196 (36%), Gaps = 44/196 (22%)
Query: 68 LVTGAGGRTGKIVYKKLQERSSQYIARGLVRT-------EESKQTIGASDDVYVGDIRDT 120
LVTGAGG G+ + + L + R L R+ E SK V GDI D
Sbjct: 6 LVTGAGGFLGQRIIRMLAQEKELQEVRTLFRSFTPKHREELSKLQTKTKVTVLEGDILDA 65
Query: 121 GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQ----VDWIGQKNQ 176
+ A QGI +I +A+ + GA P Q ++ G ++
Sbjct: 66 QCLRRACQGISVVIHTAAAIDVF------------------GAIPRQTVIDINLKGTQHL 107
Query: 177 IDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPY------- 229
+DA A V + S+ N+ + NG+ + E + + PY
Sbjct: 108 LDACIGARVPVFIYSSSVAVAGPNSYKVIIQNGS------EEENHESTWSDPYAYSKKMA 161
Query: 230 --TIIRAGGLQDKEGG 243
++ A G K+GG
Sbjct: 162 EKAVLAANGSTLKDGG 177
>GALE_METJA (Q57664) Putative UDP-glucose 4-epimerase (EC 5.1.3.2)
(Galactowaldenase) (UDP-galactose 4-epimerase)
Length = 305
Score = 34.3 bits (77), Expect = 0.41
Identities = 57/241 (23%), Positives = 93/241 (37%), Gaps = 44/241 (18%)
Query: 67 VLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPA 126
+LVTG G G + KL E + I + T +K I + DIRD +
Sbjct: 2 ILVTGGAGFIGSHIVDKLIENNYDVIILDNL-TTGNKNNINPKAEFVNADIRDK-DLDEK 59
Query: 127 IQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVK 186
I D +++ A + E+ Y ++ +G N ++ + +
Sbjct: 60 INFKDVEVVIHQAAQI-----------NVRNSVENPVYDGDINVLGTINILEMMRKYDID 108
Query: 187 QIVLVGSMGG---------TDLNNPLNSLGNGNI--LVWKRKAEQYLADSGIPYTIIR-- 233
+IV S G D N+P+N L + V + + Y GI Y I+R
Sbjct: 109 KIVFASSGGAVYGEPNYLPVDENHPINPLSPYGLSKYVGEEYIKLYNRLYGIEYAILRYS 168
Query: 234 --AGGLQDKEG--GVRELIIGKDDEILKTETRTIARP-----------DVAEVCIQALNF 278
G QD +G GV + I D++LK ++ I DVA+ + ALN+
Sbjct: 169 NVYGERQDPKGEAGVISIFI---DKMLKNQSPIIFGDGNQTRDFVYVGDVAKANLMALNW 225
Query: 279 E 279
+
Sbjct: 226 K 226
>3BH6_MOUSE (O35469) 3 beta-hydroxysteroid dehydrogenase/delta
5-->4-isomerase type VI (3Beta-HSD VI) [Includes:
3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC
1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase)
(Progesterone reductase); Steroid de
Length = 372
Score = 34.3 bits (77), Expect = 0.41
Identities = 41/154 (26%), Positives = 61/154 (38%), Gaps = 29/154 (18%)
Query: 68 LVTGAGGRTG-KIVYKKLQERSSQYIA--RGLVRTEESKQTIGASDDVYV----GDIRDT 120
LVTGAGG G +IV +QE+ + I R E +Q ++ V GDI DT
Sbjct: 6 LVTGAGGFLGQRIVQLLMQEKDLEEIRVLDKFFRPETREQFFNLDTNIKVTVLEGDILDT 65
Query: 121 GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQ----VDWIGQKNQ 176
+ A QGI +I + + + G P Q V+ G +N
Sbjct: 66 QYLRKACQGISVVIHTAAVIDV------------------TGVIPRQTILDVNLKGTQNL 107
Query: 177 IDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGN 210
++A A V + S+ N+ + NGN
Sbjct: 108 LEACIQASVPAFIFSSSVDVAGPNSYKEIILNGN 141
>3BH1_RAT (P22071) 3 beta-hydroxysteroid dehydrogenase/delta
5-->4-isomerase type I (3Beta-HSD I) [Includes:
3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC
1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase)
(Progesterone reductase); Steroid delta-
Length = 372
Score = 34.3 bits (77), Expect = 0.41
Identities = 30/80 (37%), Positives = 42/80 (52%), Gaps = 8/80 (10%)
Query: 68 LVTGAGGRTGK------IVYKKLQE-RSSQYIARGLVRTEESKQTIGASDDVYVGDIRDT 120
LVTGAGG G+ + K+LQE R+ + R + E SK A + GDI D
Sbjct: 6 LVTGAGGFVGQRIIRMLVQEKELQEVRALDKVFRPETKEEFSKLQTKAKVTMLEGDILDA 65
Query: 121 GSIAPAIQGIDALIILTSAV 140
+ A QGI +++I T+AV
Sbjct: 66 QYLRRACQGI-SVVIHTAAV 84
>3BH1_MOUSE (P24815) 3 beta-hydroxysteroid dehydrogenase/delta
5-->4-isomerase type I (3Beta-HSD I) [Includes:
3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC
1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase)
(Progesterone reductase); Steroid delt
Length = 372
Score = 34.3 bits (77), Expect = 0.41
Identities = 30/80 (37%), Positives = 41/80 (50%), Gaps = 8/80 (10%)
Query: 68 LVTGAGGRTGK------IVYKKLQE-RSSQYIARGLVRTEESKQTIGASDDVYVGDIRDT 120
LVTGAGG G+ + K+LQE R+ + R + E SK V GDI D
Sbjct: 6 LVTGAGGFVGQRIIKMLVQEKELQEVRALDKVFRPETKEEFSKLQTKTKVTVLEGDILDA 65
Query: 121 GSIAPAIQGIDALIILTSAV 140
+ A QGI +++I T+AV
Sbjct: 66 QCLRRACQGI-SVVIHTAAV 84
>GNE_ECO57 (Q8X7P7) UDP-N-acetylglucosamine 4-epimerase (EC 5.1.3.7)
(UDP-GlcNAc 4-epimerase)
Length = 331
Score = 33.9 bits (76), Expect = 0.54
Identities = 34/138 (24%), Positives = 64/138 (45%), Gaps = 24/138 (17%)
Query: 67 VLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPA 126
VL+ GA G G +L E + IA ++ + +Q+ + +GD+RD ++ A
Sbjct: 5 VLLIGASGFVGT----RLLETA---IADFNIKNLDKQQSHFYPEITQIGDVRDQQALDQA 57
Query: 127 IQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVK 186
+ G D +++L + + +PT Y++ V+ G +N + A + GVK
Sbjct: 58 LAGFDTVVLLAAE---HRDDVSPT-----SLYYD-------VNVQGTRNVLAAMEKNGVK 102
Query: 187 QIVLVGSMG--GTDLNNP 202
I+ S+ G + +NP
Sbjct: 103 NIIFTSSVAVYGLNKHNP 120
>YC39_GUITH (O78472) Hypothetical 35.9 kDa protein ycf39
Length = 314
Score = 33.5 bits (75), Expect = 0.70
Identities = 41/174 (23%), Positives = 77/174 (43%), Gaps = 32/174 (18%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQY--IARGLVRTEESKQTIGASDDVYVGDIRDTGSI 123
++LV GA G G+ + ++ + + + R L + K+ GA ++ GD+ ++
Sbjct: 2 SLLVIGATGTLGRQIVRRALDEGYEVSCLVRNLRKAYFLKEW-GA--ELLYGDLSLPETL 58
Query: 124 APAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAA 183
+ I A+I ++A P +P K E+ +D G+ ++AAK A
Sbjct: 59 PTNLTKITAIIDASTARPS-----DPYKAEK-------------IDLEGKIALVEAAKVA 100
Query: 184 GVKQIVLVGSMGGTDLNN-PLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGG 236
G+K+ V + + + PL +L K + E+YL S + YT + G
Sbjct: 101 GIKRFVFFSVLNAQNYRHLPLVNL--------KCRMEEYLQTSELEYTTFQLSG 146
>NSDL_MOUSE (Q9R1J0) NAD(P)-dependent steroid dehydrogenase (EC
1.1.1.-)
Length = 362
Score = 33.5 bits (75), Expect = 0.70
Identities = 23/98 (23%), Positives = 43/98 (43%), Gaps = 20/98 (20%)
Query: 113 YVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIG 172
++GD+ + + PA++G+ + S P + FY +V++IG
Sbjct: 70 FIGDLCNQQDLYPALKGVSTVFHCASPPPY--------SNNKELFY--------RVNFIG 113
Query: 173 QKNQIDAAKAAGVKQIVLVGSMG----GTDLNNPLNSL 206
K I+ + AGV++++L S G D+ N L
Sbjct: 114 TKTVIETCREAGVQKLILTSSASVVFEGVDIKNGTEDL 151
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,318,072
Number of Sequences: 164201
Number of extensions: 1470285
Number of successful extensions: 3976
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 3950
Number of HSP's gapped (non-prelim): 43
length of query: 313
length of database: 59,974,054
effective HSP length: 110
effective length of query: 203
effective length of database: 41,911,944
effective search space: 8508124632
effective search space used: 8508124632
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0147b.4


