

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
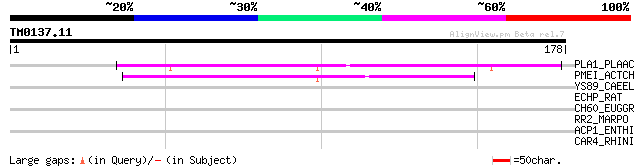
Query= TM0137.11
(178 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PLA1_PLAAC (Q8GT41) Putative invertase inhibitor precursor (Poll... 58 1e-08
PMEI_ACTCH (P83326) Pectinesterase inhibitor (Pectin methylester... 56 5e-08
YS89_CAEEL (Q09624) Hypothetical protein ZK945.9 in chromosome II 32 0.77
ECHP_RAT (P07896) Peroxisomal bifunctional enzyme (PBE) (PBFE) [... 30 3.8
CH60_EUGGR (Q39727) Chaperonin CPN60, mitochondrial precursor (H... 29 5.0
RR2_MARPO (P06354) Chloroplast 30S ribosomal protein S2 29 6.6
ACP1_ENTHI (P36184) Cysteine proteinase ACP1 precursor (EC 3.4.2... 29 6.6
CAR4_RHINI (Q03700) Rhizopuspepsin 4 precursor (EC 3.4.23.21) (A... 28 8.6
>PLA1_PLAAC (Q8GT41) Putative invertase inhibitor precursor (Pollen
allergen Pla a 1)
Length = 179
Score = 57.8 bits (138), Expect = 1e-08
Identities = 48/155 (30%), Positives = 74/155 (46%), Gaps = 13/155 (8%)
Query: 35 AQLVAKTCKNTPYPSA------CLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKI 88
A +V TCK S C++ L ADP+S +AD+ GL +I ++ + + I
Sbjct: 24 ADIVQGTCKKVAQRSPNVNYDFCVKSLGADPKSHTADLQGLGVISANLAIQHGSKIQTFI 83
Query: 89 SQLLKGGGD---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTC 145
++LK D K LN C G Y A K+ + +A K+ + A ++ A ++ TC
Sbjct: 84 GRILKSKVDPALKKYLNDCVGLY-ADAKSSVQEAIADFKSKDYASANVKMSAALDDSVTC 142
Query: 146 ESGFSSGK---SPLTSENNGMHIATEVARAIIRNL 177
E GF K SP+T EN T ++ AI + L
Sbjct: 143 EDGFKEKKGIVSPVTKENKDYVQLTAISLAITKLL 177
>PMEI_ACTCH (P83326) Pectinesterase inhibitor (Pectin methylesterase
inhibitor) (PMEI)
Length = 152
Score = 55.8 bits (133), Expect = 5e-08
Identities = 37/116 (31%), Positives = 54/116 (45%), Gaps = 4/116 (3%)
Query: 37 LVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGG 96
L+++ C T PS CLQ L++DPRS+S D+ GL +D+ QA I+ L
Sbjct: 4 LISEICPKTRNPSLCLQALESDPRSASKDLKGLGQFSIDIAQASAKQTSKIIASLTNQAT 63
Query: 97 D---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGF 149
D K +C Y + + + QA Q L +G+ + A A TCE F
Sbjct: 64 DPKLKGRYETCSENYADAIDS-LGQAKQFLTSGDYNSLNIYASAAFDGAGTCEDSF 118
>YS89_CAEEL (Q09624) Hypothetical protein ZK945.9 in chromosome II
Length = 3178
Score = 32.0 bits (71), Expect = 0.77
Identities = 24/103 (23%), Positives = 45/103 (43%), Gaps = 3/103 (2%)
Query: 60 RSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILK---ADI 116
RSS A V+ ++ +I A+ V+N+++ ++ G +LN+ N I AD+
Sbjct: 1257 RSSLATVSPISAAEQAIIDAQKADVMNQLAGIMDGSASNNSLNTSSSLLNQISSLPAADL 1316
Query: 117 PQATQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSE 159
+ Q+L + K G + T + ++ S L E
Sbjct: 1317 VEVAQSLLSNTLKIPGVGNMSSVDVLKTLQDNIATTNSELADE 1359
>ECHP_RAT (P07896) Peroxisomal bifunctional enzyme (PBE) (PBFE)
[Includes: Enoyl-CoA hydratase (EC 4.2.1.17);
3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8);
3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]
Length = 721
Score = 29.6 bits (65), Expect = 3.8
Identities = 25/71 (35%), Positives = 32/71 (44%), Gaps = 1/71 (1%)
Query: 57 ADPRSSSADVTGLAL-IMVDVIQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILKAD 115
AD SA GLAL +VD IQ VL I + GGG + AL NA +
Sbjct: 60 ADIHGFSAFTPGLALGSLVDEIQRYQKPVLAAIQGVALGGGLELALGCHYRIANAKARVG 119
Query: 116 IPQATQALKTG 126
+P+ T + G
Sbjct: 120 LPEVTLGILPG 130
>CH60_EUGGR (Q39727) Chaperonin CPN60, mitochondrial precursor (HSP
60)
Length = 569
Score = 29.3 bits (64), Expect = 5.0
Identities = 31/124 (25%), Positives = 51/124 (41%), Gaps = 12/124 (9%)
Query: 47 YPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNK-------ISQLLKGGGDKP 99
Y S+ L L ADP + TG+ ++M + V N I QLL +
Sbjct: 435 YASSALAELAADPTLTEDQKTGVRIVMSAIKLPAITIVKNAGGEGAVVIHQLLAEKKMQQ 494
Query: 100 ALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSE 159
++ QG+Y + +A I + +KT D + AG+ T E+ + +P +
Sbjct: 495 GYDAQQGKYVNMFEAGIIDPAKVVKTA----LVDAASVAGLMITT-EAAITDIPAPAPAA 549
Query: 160 NNGM 163
GM
Sbjct: 550 GGGM 553
>RR2_MARPO (P06354) Chloroplast 30S ribosomal protein S2
Length = 235
Score = 28.9 bits (63), Expect = 6.6
Identities = 20/70 (28%), Positives = 30/70 (42%), Gaps = 16/70 (22%)
Query: 42 CKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGDKPAL 101
C P+ CL DP D+T + + D +A +LNK++ L
Sbjct: 180 CITLGIPTICLVDTDCDP-----DMTDIPIPANDDARASIRWILNKLT-----------L 223
Query: 102 NSCQGRYNAI 111
C+GRYN+I
Sbjct: 224 AICEGRYNSI 233
>ACP1_ENTHI (P36184) Cysteine proteinase ACP1 precursor (EC
3.4.22.-)
Length = 308
Score = 28.9 bits (63), Expect = 6.6
Identities = 12/39 (30%), Positives = 18/39 (45%)
Query: 130 FAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATE 168
F+E + D N CE G S ENNG+ + ++
Sbjct: 138 FSEQQLVDCDASDNGCEGGHPSNSLKFIQENNGLGLESD 176
>CAR4_RHINI (Q03700) Rhizopuspepsin 4 precursor (EC 3.4.23.21)
(Aspartate protease)
Length = 398
Score = 28.5 bits (62), Expect = 8.6
Identities = 16/55 (29%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Query: 117 PQATQALKTGNPKFAEDGVADAGVEANTCESG--FSSGKSPLTSENNGMHIATEV 169
P A +A++ N K+A + + +++C S SSG P+T + N + EV
Sbjct: 40 PNAKRAIEKANAKYARFRSSSSSSSSSSCGSAGTESSGSVPVTDDGNDIEYYGEV 94
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,960,398
Number of Sequences: 164201
Number of extensions: 712226
Number of successful extensions: 1761
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1758
Number of HSP's gapped (non-prelim): 8
length of query: 178
length of database: 59,974,054
effective HSP length: 103
effective length of query: 75
effective length of database: 43,061,351
effective search space: 3229601325
effective search space used: 3229601325
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0137.11


