

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.9
(645 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
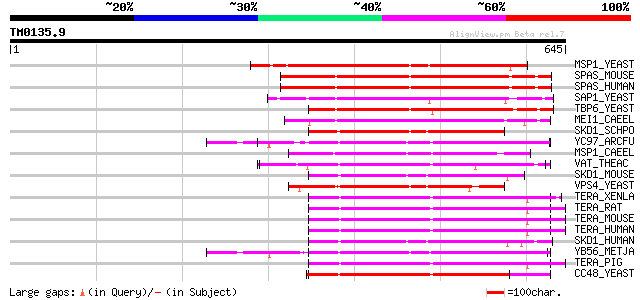
Score E
Sequences producing significant alignments: (bits) Value
MSP1_YEAST (P28737) MSP1 protein (TAT-binding homolog 4) 246 1e-64
SPAS_MOUSE (Q9QYY8) Spastin 238 3e-62
SPAS_HUMAN (Q9UBP0) Spastin 238 4e-62
SAP1_YEAST (P39955) SAP1 protein 227 8e-59
TBP6_YEAST (P40328) Probable 26S protease subunit YTA6 (TAT-bind... 211 4e-54
MEI1_CAEEL (P34808) Meiotic spindle formation protein mei-1 206 2e-52
SKD1_SCHPO (Q09803) Suppressor protein of bem1/bed5 double mutants 201 4e-51
YC97_ARCFU (O28972) Cell division cycle protein 48 homolog AF1297 199 2e-50
MSP1_CAEEL (P54815) MSP1 protein homolog 198 3e-50
VAT_THEAC (O05209) VCP-like ATPase 196 2e-49
SKD1_MOUSE (P46467) SKD1 protein (Vacuolar sorting protein 4b) 190 9e-48
VPS4_YEAST (P52917) Vacuolar protein sorting-associated protein ... 190 1e-47
TERA_XENLA (P23787) Transitional endoplasmic reticulum ATPase (T... 187 1e-46
TERA_RAT (P46462) Transitional endoplasmic reticulum ATPase (TER... 186 2e-46
TERA_MOUSE (Q01853) Transitional endoplasmic reticulum ATPase (T... 186 2e-46
TERA_HUMAN (P55072) Transitional endoplasmic reticulum ATPase (T... 186 2e-46
SKD1_HUMAN (O75351) SKD1 protein (Vacuolar sorting protein 4b) 186 2e-46
YB56_METJA (Q58556) Cell division cycle protein 48 homolog MJ1156 184 5e-46
TERA_PIG (P03974) Transitional endoplasmic reticulum ATPase (TER... 184 5e-46
CC48_YEAST (P25694) Cell division control protein 48 183 1e-45
>MSP1_YEAST (P28737) MSP1 protein (TAT-binding homolog 4)
Length = 362
Score = 246 bits (629), Expect = 1e-64
Identities = 140/330 (42%), Positives = 205/330 (61%), Gaps = 16/330 (4%)
Query: 281 HYLSSCLLPCVKGDKLC-LPRESLEIAISRLKGVETMSRQPSQSLKNLAKDEFESNFISA 339
+YL S LL V+ L RES A L+ + + R P+ L + D +E +S+
Sbjct: 24 YYLVSRLLNDVESGPLSGKSRESK--AKQSLQWEKLVKRSPA--LAEVTLDAYERTILSS 79
Query: 340 VVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGK 399
+V P EI + F DIG L+ + L+E VI P+ PE++++ LL+ G+LL+GPPG GK
Sbjct: 80 IVTPDEINITFQDIGGLDPLISDLHESVIYPLMMPEVYSNSPLLQAPSGVLLYGPPGCGK 139
Query: 400 TLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLG 459
T++AKALA E+GANFIS+ S++ KW+G++ K+ A+FS A+KL P IIF+DE+DS L
Sbjct: 140 TMLAKALAKESGANFISIRMSSIMDKWYGESNKIVDAMFSLANKLQPCIIFIDEIDSFLR 199
Query: 460 ARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVD 519
R +HE T ++ EFM WDGL + N R++I+GATNR D+DDA +RRLP+R V
Sbjct: 200 ERSST-DHEVTATLKAEFMTLWDGLLN--NGRVMIIGATNRINDIDDAFLRRLPKRFLVS 256
Query: 520 LPDADNRKKILRIILKHENLDPD-FQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEE 578
LP +D R KIL ++LK LD D F +A+ T G+SGSDLK LC AA +E +++
Sbjct: 257 LPGSDQRYKILSVLLKDTKLDEDEFDLQLIADNTKGFSGSDLKELCREAALDAAKEYIKQ 316
Query: 579 EK-------AGDNNITNVALRPLNLDDFVQ 601
++ N+ +++ +RPL DF +
Sbjct: 317 KRQLIDSGTIDVNDTSSLKIRPLKTKDFTK 346
>SPAS_MOUSE (Q9QYY8) Spastin
Length = 614
Score = 238 bits (608), Expect = 3e-62
Identities = 136/317 (42%), Positives = 202/317 (62%), Gaps = 10/317 (3%)
Query: 315 TMSRQPSQSLKNLAK-DEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRR 373
T + + + LKN D +N I + VKFDDI E K+AL E+VILP R
Sbjct: 303 TTAVRKKKDLKNFRNVDSNLANLIMNEIVDNGTAVKFDDIAGQELAKQALQEIVILPSLR 362
Query: 374 PELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKL 433
PELFT L P +G+LLFGPPG GKT++AKA+A E+ A F ++S ++LTSK+ G+ EKL
Sbjct: 363 PELFT--GLRAPARGLLLFGPPGNGKTMLAKAVAAESNATFFNISAASLTSKYVGEGEKL 420
Query: 434 TKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRIL 493
+ALF+ A +L P IIF+DEVDSLL R EH+A+RR++ EF+ +DG++S + R+L
Sbjct: 421 VRALFAVARELQPSIIFIDEVDSLLCERREG-EHDASRRLKTEFLIEFDGVQSAGDDRVL 479
Query: 494 ILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILR-IILKHENLDPDFQFDKLANLT 552
++GATNRP +LD+AV+RR +R+YV LP+ + R +L+ ++ K + + +LA +T
Sbjct: 480 VMGATNRPQELDEAVLRRFIKRVYVSLPNEETRLLLLKNLLCKQGSPLTQKELAQLARMT 539
Query: 553 DGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAY 612
DGYSGSDL L AA P++EL E+ N++ +R + L DF +S K+ SV+
Sbjct: 540 DGYSGSDLTALAKDAALGPIRELKPEQV---KNMSASEMRNIRLSDFTESLKKIKRSVS- 595
Query: 613 DAVSVNELRKWNEMYGE 629
++ +WN+ +G+
Sbjct: 596 -PQTLEAYIRWNKDFGD 611
>SPAS_HUMAN (Q9UBP0) Spastin
Length = 616
Score = 238 bits (607), Expect = 4e-62
Identities = 135/317 (42%), Positives = 202/317 (63%), Gaps = 10/317 (3%)
Query: 315 TMSRQPSQSLKNLAK-DEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRR 373
T + + + LKN D +N I + VKFDDI + K+AL E+VILP R
Sbjct: 305 TTATRKKKDLKNFRNVDSNLANLIMNEIVDNGTAVKFDDIAGQDLAKQALQEIVILPSLR 364
Query: 374 PELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKL 433
PELFT L P +G+LLFGPPG GKT++AKA+A E+ A F ++S ++LTSK+ G+ EKL
Sbjct: 365 PELFT--GLRAPARGLLLFGPPGNGKTMLAKAVAAESNATFFNISAASLTSKYVGEGEKL 422
Query: 434 TKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRIL 493
+ALF+ A +L P IIF+DEVDSLL R EH+A+RR++ EF+ +DG++S + R+L
Sbjct: 423 VRALFAVARELQPSIIFIDEVDSLLCERREG-EHDASRRLKTEFLIEFDGVQSAGDDRVL 481
Query: 494 ILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILR-IILKHENLDPDFQFDKLANLT 552
++GATNRP +LD+AV+RR +R+YV LP+ + R +L+ ++ K + + +LA +T
Sbjct: 482 VMGATNRPQELDEAVLRRFIKRVYVSLPNEETRLLLLKNLLCKQGSPLTQKELAQLARMT 541
Query: 553 DGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAY 612
DGYSGSDL L AA P++EL E+ N++ +R + L DF +S K+ SV+
Sbjct: 542 DGYSGSDLTALAKDAALGPIRELKPEQV---KNMSASEMRNIRLSDFTESLKKIKRSVS- 597
Query: 613 DAVSVNELRKWNEMYGE 629
++ +WN+ +G+
Sbjct: 598 -PQTLEAYIRWNKDFGD 613
>SAP1_YEAST (P39955) SAP1 protein
Length = 897
Score = 227 bits (578), Expect = 8e-59
Identities = 138/356 (38%), Positives = 206/356 (57%), Gaps = 39/356 (10%)
Query: 300 RESLEIAISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDV 359
RE EI L+ E + + SL+ + + + F VV E V +DDI LE
Sbjct: 558 REEPEIDKKVLR--EILEDEIIDSLQGVDRQAAKQIFAEIVVHGDE--VHWDDIAGLESA 613
Query: 360 KKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSG 419
K +L E V+ P RP+LF L PV+G+LLFGPPGTGKT++A+A+ATE+ + F S+S
Sbjct: 614 KYSLKEAVVYPFLRPDLFR--GLREPVRGMLLFGPPGTGKTMLARAVATESHSTFFSISA 671
Query: 420 STLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMA 479
S+LTSK+ G++EKL +ALF+ A KL+P IIFVDE+DS++G+R E+E++RR++NEF+
Sbjct: 672 SSLTSKYLGESEKLVRALFAIAKKLSPSIIFVDEIDSIMGSRNNENENESSRRIKNEFLV 731
Query: 480 AWDGLRS------------------KENQRILILGATNRPFDLDDAVIRRLPRRIYVDLP 521
W L S +++ R+L+L ATN P+ +D+A RR RR Y+ LP
Sbjct: 732 QWSSLSSAAAGSNKSNTNNSDTNGDEDDTRVLVLAATNLPWSIDEAARRRFVRRQYIPLP 791
Query: 522 DADNRKKILRIILKHE-NLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQE----LL 576
+ R + +L H+ + + FD+L +T+GYSGSD+ +L AA P+++ LL
Sbjct: 792 EDQTRHVQFKKLLSHQKHTLTESDFDELVKITEGYSGSDITSLAKDAAMGPLRDLGDKLL 851
Query: 577 EEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGT 632
E E+ +RP+ L DF S + PSV+ D + E KW +G G+
Sbjct: 852 ETERE--------MIRPIGLVDFKNSLVYIKPSVSQDGLVKYE--KWASQFGSSGS 897
>TBP6_YEAST (P40328) Probable 26S protease subunit YTA6 (TAT-binding
homolog 6)
Length = 754
Score = 211 bits (538), Expect = 4e-54
Identities = 117/296 (39%), Positives = 183/296 (61%), Gaps = 20/296 (6%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DI L + K +L E V+ P RP+LF L PV+G+LLFGPPGTGKT++AKA+A
Sbjct: 468 VYWEDIAGLRNAKNSLKEAVVYPFLRPDLFK--GLREPVRGMLLFGPPGTGKTMIAKAVA 525
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
TE+ + F SVS S+L SK+ G++EKL +ALF A KL+P IIF+DE+DS+L AR E+
Sbjct: 526 TESNSTFFSVSASSLLSKYLGESEKLVRALFYMAKKLSPSIIFIDEIDSMLTARSDN-EN 584
Query: 468 EATRRMRNEFMAAWDGLRSKENQ----------RILILGATNRPFDLDDAVIRRLPRRIY 517
E++RR++ E + W L S Q R+L+LGATN P+ +DDA RR R++Y
Sbjct: 585 ESSRRIKTELLIQWSSLSSATAQSEDRNNTLDSRVLVLGATNLPWAIDDAARRRFSRKLY 644
Query: 518 VDLPDADNR-KKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELL 576
+ LPD + R + R++ K +N D ++ + +T+G+SGSDL +L AA P+++L
Sbjct: 645 IPLPDYETRLYHLKRLMAKQKNSLQDLDYELITEMTEGFSGSDLTSLAKEAAMEPIRDLG 704
Query: 577 EEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGT 632
++ D + +R + + DF + + SV+ + S+ + +W+ +G G+
Sbjct: 705 DKLMFADFD----KIRGIEIKDFQNALLTIKKSVSSE--SLQKYEEWSSKFGSNGS 754
>MEI1_CAEEL (P34808) Meiotic spindle formation protein mei-1
Length = 472
Score = 206 bits (524), Expect = 2e-52
Identities = 120/326 (36%), Positives = 194/326 (58%), Gaps = 23/326 (7%)
Query: 320 PSQSL--KNLAKDEFESNFISAVVPPGEIG---------VKFDDIGALEDVKKALNELVI 368
P+Q + +N A D F+++ A + G + DDI + DVK+ L+E V
Sbjct: 151 PTQGILPQNSAGDSFDASAYDAYIVQAVRGTMATNTENTMSLDDIIGMHDVKQVLHEAVT 210
Query: 369 LPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFG 428
LP+ PE F L P K ++L GPPGTGKTL+A+A+A+E+ + F +VS + L+SKW G
Sbjct: 211 LPLLVPEFFQ--GLRSPWKAMVLAGPPGTGKTLIARAIASESSSTFFTVSSTDLSSKWRG 268
Query: 429 DAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSK- 487
D+EK+ + LF A AP IIF+DE+D+L G RG + EHEA+RR+++EF+ DG ++K
Sbjct: 269 DSEKIVRLLFELARFYAPSIIFIDEIDTLGGQRGNSGEHEASRRVKSEFLVQMDGSQNKF 328
Query: 488 ENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDK 547
+++R+ +L ATN P++LD+A+ RR +RI++ LPD D RKK++ ++ + +D
Sbjct: 329 DSRRVFVLAATNIPWELDEALRRRFEKRIFIPLPDIDARKKLIEKSMEGTPKSDEINYDD 388
Query: 548 LANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNLD-----DFVQS 602
LA T+G+SG+D+ +LC TAA ++ + K+ A+ L + DF +
Sbjct: 389 LAARTEGFSGADVVSLCRTAAINVLRRY--DTKSLRGGELTAAMESLKAELVRNIDFEAA 446
Query: 603 KSKVGPSVAYDAVSVNELRKWNEMYG 628
V PS D ++ + ++W + +G
Sbjct: 447 LQAVSPSAGPD--TMLKCKEWCDSFG 470
>SKD1_SCHPO (Q09803) Suppressor protein of bem1/bed5 double mutants
Length = 432
Score = 201 bits (512), Expect = 4e-51
Identities = 105/229 (45%), Positives = 155/229 (66%), Gaps = 5/229 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V++DDI LE+ K+AL E V+LP++ P+LF+HG +P GILL+GPPGTGK+ +AKA+A
Sbjct: 126 VRWDDIAGLENAKEALKETVLLPIKLPQLFSHGR--KPWSGILLYGPPGTGKSYLAKAVA 183
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
TEAG+ F S+S S L SKW G++E+L + LF A + P IIF+DE+DSL G+R E
Sbjct: 184 TEAGSTFFSISSSDLVSKWMGESERLVRQLFEMAREQKPSIIFIDEIDSLCGSRSEG-ES 242
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRK 527
E++RR++ EF+ +G+ K+ +L+LGATN P+ LD A+ RR +RIY+ LP+A R
Sbjct: 243 ESSRRIKTEFLVQMNGV-GKDESGVLVLGATNIPWTLDSAIRRRFEKRIYIPLPNAHARA 301
Query: 528 KILRI-ILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQEL 575
++ + + K + F +LA +TDGYSGSD+ + A PV+ +
Sbjct: 302 RMFELNVGKIPSELTSQDFKELAKMTDGYSGSDISIVVRDAIMEPVRRI 350
>YC97_ARCFU (O28972) Cell division cycle protein 48 homolog AF1297
Length = 733
Score = 199 bits (506), Expect = 2e-50
Identities = 134/406 (33%), Positives = 215/406 (52%), Gaps = 22/406 (5%)
Query: 229 DRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLL 288
DR+I I + +++LE H +D +++ AE G+ + C
Sbjct: 342 DREIEIGVPDKEGRKEILEIHTRKMPLAEDVDLEEL------AELTNGFVGADLEALCKE 395
Query: 289 PCVKGDKLCLPR---ESLEIAISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGE 345
+ + LP E+ EI ++ ++ ++LKN+ + VP
Sbjct: 396 AAMHALRRVLPEIDIEAEEIPAEVIENLKVTREDFMEALKNIEPSAMREVLVE--VP--- 450
Query: 346 IGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKA 405
VK++DIG LE K+ L E V P++ PE+F N+ +P +GILLFGPPGTGKTL+AKA
Sbjct: 451 -NVKWEDIGGLEHAKQELMEAVEWPLKYPEVFRAANI-KPPRGILLFGPPGTGKTLLAKA 508
Query: 406 LATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAF 465
+A E+ ANFISV G L SKW G++EK + +F A ++AP +IF DE+DSL RGG
Sbjct: 509 VANESNANFISVKGPELLSKWVGESEKHVREMFRKARQVAPCVIFFDEIDSLAPRRGGIG 568
Query: 466 EHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
+ T R+ ++ + DGL +E + ++++ ATNRP +D A++R RL R IY+ PD
Sbjct: 569 DSHVTERVVSQLLTELDGL--EELKDVVVIAATNRPDMIDPALLRPGRLERHIYIPPPDK 626
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGD 583
R +I +I L+ + L D ++LA T+GYSG+D++ +C A ++EL++ +
Sbjct: 627 KARVEIFKIHLRGKPLADDVNIEELAEKTEGYSGADIEAVCREAGMLAIRELIKPGMTRE 686
Query: 584 NNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNE--LRKWNEMY 627
+ F ++ KV PS+ + V E + ++ MY
Sbjct: 687 EAKEAAKKLKITKKHFEEALKKVRPSLTKEDVEKYEKLIEDFHRMY 732
Score = 181 bits (459), Expect = 5e-45
Identities = 116/348 (33%), Positives = 191/348 (54%), Gaps = 21/348 (6%)
Query: 289 PCVKGDKLCLP----RESLEIAISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPG 344
P +KG K+ + + I +R GV ++R + LK +E + VP
Sbjct: 125 PVIKGQKIRVEVFGHTLTFVITATRPSGVVVVTRNTAIELKEKPAEE-----VKRAVPD- 178
Query: 345 EIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAK 404
V ++DIG L+ + + E++ LP++ PELF + P KG+LL+GPPGTGKTL+AK
Sbjct: 179 ---VTYEDIGGLKRELRLVREMIELPLKHPELFQRLGI-EPPKGVLLYGPPGTGKTLIAK 234
Query: 405 ALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGA 464
A+A E A+FI +SG + SK++G++E+ + +F A + AP IIF+DE+DS+ R
Sbjct: 235 AVANEVDAHFIPISGPEIMSKYYGESEQRLREIFEEAKENAPSIIFIDEIDSIAPKR-EE 293
Query: 465 FEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPD 522
E RR+ + +A DGL ++ + ++++ ATNRP +D A+ R R R I + +PD
Sbjct: 294 VTGEVERRVVAQLLALMDGLEARGD--VIVIAATNRPDAIDPALRRPGRFDREIEIGVPD 351
Query: 523 ADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAG 582
+ RK+IL I + L D ++LA LT+G+ G+DL+ LC AA ++ +L E
Sbjct: 352 KEGRKEILEIHTRKMPLAEDVDLEELAELTNGFVGADLEALCKEAAMHALRRVLPEIDIE 411
Query: 583 DNNITNVALRPLNL--DDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I + L + +DF+++ + PS + + KW ++ G
Sbjct: 412 AEEIPAEVIENLKVTREDFMEALKNIEPSAMREVLVEVPNVKWEDIGG 459
>MSP1_CAEEL (P54815) MSP1 protein homolog
Length = 342
Score = 198 bits (504), Expect = 3e-50
Identities = 113/282 (40%), Positives = 168/282 (59%), Gaps = 11/282 (3%)
Query: 325 KNLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLR 384
+ + E E + V ++G +D+IG E++ L + +ILP+R + +LL
Sbjct: 57 RQIELSEHEIRIATQFVGGEDVGADWDEIGGCEELVAELKDRIILPLRFASQ-SGSHLLS 115
Query: 385 PVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKL 444
P +GILL+GPPG GKTL+AKA+A AG FI++ S LT KW+G+++KL A+FS A K
Sbjct: 116 PPRGILLYGPPGCGKTLLAKAVARAAGCRFINLQVSNLTDKWYGESQKLAAAVFSVAQKF 175
Query: 445 APVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDL 504
P IIF+DE+DS L R + +HE+T M+ +FM WDG S +Q I+++GATNRP D+
Sbjct: 176 QPTIIFIDEIDSFLRDR-QSHDHESTAMMKAQFMTLWDGFSSSGDQ-IIVMGATNRPRDV 233
Query: 505 DDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLC 564
D A++RR+ R V +P+A R +IL +IL++E ++ ++A +G SGSDLK +C
Sbjct: 234 DAAILRRMTARFQVPVPNAKQRSQILNVILRNEKINNTVNLGEIAQAAEGLSGSDLKEVC 293
Query: 565 ITAAYRPVQELLEEEKAGDNNITNV-ALRPLNLDDFVQSKSK 605
A LL KA + +V L PL DF + K
Sbjct: 294 RLA-------LLARAKATVASGGSVNQLLPLEQSDFESAVHK 328
>VAT_THEAC (O05209) VCP-like ATPase
Length = 745
Score = 196 bits (497), Expect = 2e-49
Identities = 122/346 (35%), Positives = 197/346 (56%), Gaps = 21/346 (6%)
Query: 291 VKGDKLCLPRESLEIAISR-LKGVETMSRQPSQSLKNLA--KDEFESNFISAVVPPG--- 344
V D L RES A+ R L ++ P++ L+ + +D+F+ N + ++ P
Sbjct: 398 VGADLAALVRESAMNALRRYLPEIDLDKPIPTEILEKMVVTEDDFK-NALKSIEPSSLRE 456
Query: 345 ---EI-GVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKT 400
E+ V +DDIG LEDVK+ + E V LP+ +P++F + RP KG LL+GPPG GKT
Sbjct: 457 VMVEVPNVHWDDIGGLEDVKREIKETVELPLLKPDVFKRLGI-RPSKGFLLYGPPGVGKT 515
Query: 401 LMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGA 460
L+AKA+ATE+ ANFIS+ G + SKW G++EK + +F A ++AP I+F+DE+DS+
Sbjct: 516 LLAKAVATESNANFISIKGPEVLSKWVGESEKAIREIFKKAKQVAPAIVFLDEIDSIAPR 575
Query: 461 RGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYV 518
RG + T R+ N+ + + DG+ ++++GATNRP +D A++R R + IY+
Sbjct: 576 RGTTSDSGVTERIVNQLLTSLDGIEVMNG--VVVIGATNRPDIMDPALLRAGRFDKLIYI 633
Query: 519 DLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEE 578
PD + R IL++ K+ L PD + +A T+GY G+DL+NLC A +E +
Sbjct: 634 PPPDKEARLSILKVHTKNMPLAPDVDLNDIAQRTEGYVGADLENLCREAGMNAYRENPDA 693
Query: 579 EKAGDNNITNV--ALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRK 622
N + +RP ++ ++ + ++ + SV+E RK
Sbjct: 694 TSVSQKNFLDALKTIRPSVDEEVIKFYRTLSETM---SKSVSERRK 736
Score = 155 bits (393), Expect = 2e-37
Identities = 110/351 (31%), Positives = 189/351 (53%), Gaps = 18/351 (5%)
Query: 289 PCVKGDKLCLPRESLEIAISRL-KGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGEIG 347
P ++ D + +P +L L K V+T+ + + K E S V+ E+
Sbjct: 128 PMLEQDNISVPGLTLAGQTGLLFKVVKTLPSKVPVEIGEETKIEIREEPASEVLE--EVS 185
Query: 348 -VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKAL 406
+ ++DIG L + + E++ LP++ PELF + P KG++L+GPPGTGKTL+A+A+
Sbjct: 186 RISYEDIGGLSEQLGKIREMIELPLKHPELFERLGITPP-KGVILYGPPGTGKTLIARAV 244
Query: 407 ATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFE 466
A E+GANF+S++G + SK++G +E+ + +FS A + AP IIF+DE+DS+ R +
Sbjct: 245 ANESGANFLSINGPEIMSKYYGQSEQKLREIFSKAEETAPSIIFIDEIDSIAPKR-EEVQ 303
Query: 467 HEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDAD 524
E RR+ + + DG+ KE ++++GATNR +D A+ R R R I + +PD +
Sbjct: 304 GEVERRVVAQLLTLMDGM--KERGHVIVIGATNRIDAIDPALRRPGRFDREIEIGVPDRN 361
Query: 525 NRKKILRIILKHENL-----DPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE 579
RK+IL I ++ L + + +++A+ T G+ G+DL L +A ++ L E
Sbjct: 362 GRKEILMIHTRNMPLGMSEEEKNKFLEEMADYTYGFVGADLAALVRESAMNALRRYLPEI 421
Query: 580 KAGDNNITNVALRPLNL--DDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
D I L + + DDF + + PS + + W+++ G
Sbjct: 422 DL-DKPIPTEILEKMVVTEDDFKNALKSIEPSSLREVMVEVPNVHWDDIGG 471
>SKD1_MOUSE (P46467) SKD1 protein (Vacuolar sorting protein 4b)
Length = 444
Score = 190 bits (483), Expect = 9e-48
Identities = 115/262 (43%), Positives = 157/262 (59%), Gaps = 15/262 (5%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
VK+ D+ LE K+AL E VILP++ P LFT P +GILLFGPPGTGK+ +AKA+A
Sbjct: 131 VKWSDVAGLEGAKEALKEAVILPIKFPHLFTGKRT--PWRGILLFGPPGTGKSYLAKAVA 188
Query: 408 TEAG-ANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFE 466
TEA + F S+S S L SKW G++EKL K LF A + P IIF+DE+DSL G+R E
Sbjct: 189 TEANNSTFFSISSSDLVSKWLGESEKLVKNLFQLARENKPSIIFIDEIDSLCGSR-SENE 247
Query: 467 HEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNR 526
EA RR++ EF+ G+ +N IL+LGATN P+ LD A+ RR +RIY+ LP+A R
Sbjct: 248 SEAARRIKTEFLVQMQGV-GVDNDGILVLGATNIPWVLDSAIRRRFEKRIYIPLPEAHAR 306
Query: 527 KKILRIIL-KHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLE-------- 577
+ R+ L +N + F +L TDGYSG+D+ + A +PV+++
Sbjct: 307 AAMFRLHLGSTQNSLTEADFQELGRKTDGYSGADISIIVRDALMQPVRKVQSATHFKKVR 366
Query: 578 -EEKAGDNNITNVALRPLNLDD 598
+A N I N L P + D
Sbjct: 367 GPSRADPNCIVNDLLTPCSPGD 388
>VPS4_YEAST (P52917) Vacuolar protein sorting-associated protein
VPS4 (END13 protein)
Length = 437
Score = 190 bits (482), Expect = 1e-47
Identities = 112/268 (41%), Positives = 164/268 (60%), Gaps = 27/268 (10%)
Query: 325 KNLAKDEFESN----------FISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRP 374
K ++++E E N +S+ + + VK++D+ LE K+AL E VILP++ P
Sbjct: 97 KKISQEEGEDNGGEDNKKLRGALSSAILSEKPNVKWEDVAGLEGAKEALKEAVILPVKFP 156
Query: 375 ELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLT 434
LF GN +P GILL+GPPGTGK+ +AKA+ATEA + F SVS S L SKW G++EKL
Sbjct: 157 HLFK-GNR-KPTSGILLYGPPGTGKSYLAKAVATEANSTFFSVSSSDLVSKWMGESEKLV 214
Query: 435 KALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILI 494
K LF+ A + P IIF+DEVD+L G RG E EA+RR++ E + +G+ ++Q +L+
Sbjct: 215 KQLFAMARENKPSIIFIDEVDALTGTRGEG-ESEASRRIKTELLVQMNGV-GNDSQGVLV 272
Query: 495 LGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRI-------ILKHENLDPDFQFDK 547
LGATN P+ LD A+ RR RRIY+ LPD R + I +L E+ +
Sbjct: 273 LGATNIPWQLDSAIRRRFERRIYIPLPDLAARTTMFEINVGDTPCVLTKED------YRT 326
Query: 548 LANLTDGYSGSDLKNLCITAAYRPVQEL 575
L +T+GYSGSD+ + A +P++++
Sbjct: 327 LGAMTEGYSGSDIAVVVKDALMQPIRKI 354
>TERA_XENLA (P23787) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
Length = 805
Score = 187 bits (474), Expect = 1e-46
Identities = 116/303 (38%), Positives = 177/303 (58%), Gaps = 16/303 (5%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG LEDVK+ L ELV P+ P+ F + P KG+L +GPPG GKTL+AKA+A
Sbjct: 474 VTWEDIGGLEDVKRELQELVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 532
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E ANFIS+ G L + WFG++E + +F A + AP ++F DE+DS+ ARGG
Sbjct: 533 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 592
Query: 468 --EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
A R+ N+ + DG+ K+N + I+GATNRP +D A++R RL + IY+ LPD
Sbjct: 593 GGGAADRVINQILTEMDGMSIKKN--VFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 650
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE-KAG 582
+R IL+ L+ + D D LA +T+G+SG+DL +C A ++E +E E +
Sbjct: 651 KSRMAILKANLRKSPVAKDVDVDFLAKMTNGFSGADLTEICQRACKLAIRESIENEIRRE 710
Query: 583 DNNITNVALRPLNLDDFV----QSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPF 638
+ TN + + DD V + + +A +VS N++RK+ EM+ + +++
Sbjct: 711 RDRQTNPSAMEVEEDDPVPEIRRDHFEEAMRLARRSVSDNDIRKY-EMFAQTLQQSR--- 766
Query: 639 GFG 641
GFG
Sbjct: 767 GFG 769
Score = 163 bits (412), Expect = 1e-39
Identities = 100/285 (35%), Positives = 156/285 (54%), Gaps = 8/285 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V +DDIG + E+V LP+R P LF + +P +GILL+GPPGTGKTL+A+A+A
Sbjct: 201 VGYDDIGGCRKQLAQIKEMVELPLRHPALFKAIGV-KPPRGILLYGPPGTGKTLIARAVA 259
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E GA F ++G + SK G++E + F A K AP IIF+DE+D++ R
Sbjct: 260 NETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREKT-HG 318
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ ++ + DGL K+ ++++ ATNRP +D A+ R R R + + +PD+
Sbjct: 319 EVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIPDSTG 376
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
R +IL+I K+ L D +++AN T G+ G+DL LC AA + +++ ++ D
Sbjct: 377 RLEILQIHTKNMKLSDDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDLEDET 436
Query: 586 ITNVALRPL--NLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I + L +DDF S+ PS + V W ++ G
Sbjct: 437 IDAEVMNSLAVTMDDFRWGLSQSNPSALRETVVEVPQVTWEDIGG 481
>TERA_RAT (P46462) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
(Valosin-containing protein) (VCP)
Length = 806
Score = 186 bits (471), Expect = 2e-46
Identities = 117/309 (37%), Positives = 176/309 (56%), Gaps = 15/309 (4%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG LEDVK+ L ELV P+ P+ F + P KG+L +GPPG GKTL+AKA+A
Sbjct: 474 VTWEDIGGLEDVKRELQELVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 532
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E ANFIS+ G L + WFG++E + +F A + AP ++F DE+DS+ ARGG
Sbjct: 533 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 592
Query: 468 --EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
A R+ N+ + DG+ +K+N + I+GATNRP +D A++R RL + IY+ LPD
Sbjct: 593 GGGAADRVINQILTEMDGMSTKKN--VFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 650
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE-KAG 582
+R IL+ L+ + D + LA +T+G+SG+DL +C A ++E +E E +
Sbjct: 651 KSRVAILKANLRKSPVAKDVDLEFLAKMTNGFSGADLTEICQRACKLAIRESIESEIRRE 710
Query: 583 DNNITNVALRPLNLDDFV----QSKSKVGPSVAYDAVSVNELRKWNEMYGE--GGTRTKS 636
TN + + DD V + + A +VS N++RK+ EM+ + +R
Sbjct: 711 RERQTNPSAMEVEEDDPVPEIRRDHFEEAMRFARRSVSDNDIRKY-EMFAQTLQQSRGFG 769
Query: 637 PFGFGQNNQ 645
F F NQ
Sbjct: 770 SFRFPSGNQ 778
Score = 164 bits (414), Expect = 9e-40
Identities = 101/285 (35%), Positives = 157/285 (54%), Gaps = 8/285 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V +DDIG + E+V LP+R P LF + +P +GILL+GPPGTGKTL+A+A+A
Sbjct: 201 VGYDDIGGCRKQLAQIKEMVELPLRHPALFKAIGV-KPPRGILLYGPPGTGKTLIARAVA 259
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E GA F ++G + SK G++E + F A K AP IIF+DE+D++ R
Sbjct: 260 NETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREKT-HG 318
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ ++ + DGL K+ ++++ ATNRP +D A+ R R R + + +PDA
Sbjct: 319 EVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIPDATG 376
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
R +IL+I K+ L D +++AN T G+ G+DL LC AA + +++ ++ D
Sbjct: 377 RLEILQIHTKNMKLADDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDLEDET 436
Query: 586 ITNVALRPL--NLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I + L +DDF + S+ PS + V W ++ G
Sbjct: 437 IDAEVMNSLAVTMDDFRWALSQSNPSALRETVVEVPQVTWEDIGG 481
>TERA_MOUSE (Q01853) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
(Valosin-containing protein) (VCP)
Length = 806
Score = 186 bits (471), Expect = 2e-46
Identities = 117/309 (37%), Positives = 176/309 (56%), Gaps = 15/309 (4%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG LEDVK+ L ELV P+ P+ F + P KG+L +GPPG GKTL+AKA+A
Sbjct: 474 VTWEDIGGLEDVKRELQELVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 532
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E ANFIS+ G L + WFG++E + +F A + AP ++F DE+DS+ ARGG
Sbjct: 533 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 592
Query: 468 --EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
A R+ N+ + DG+ +K+N + I+GATNRP +D A++R RL + IY+ LPD
Sbjct: 593 GGGAADRVINQILTEMDGMSTKKN--VFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 650
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE-KAG 582
+R IL+ L+ + D + LA +T+G+SG+DL +C A ++E +E E +
Sbjct: 651 KSRVAILKANLRKSPVAKDVDLEFLAKMTNGFSGADLTEICQRACKLAIRESIESEIRRE 710
Query: 583 DNNITNVALRPLNLDDFV----QSKSKVGPSVAYDAVSVNELRKWNEMYGE--GGTRTKS 636
TN + + DD V + + A +VS N++RK+ EM+ + +R
Sbjct: 711 RERQTNPSAMEVEEDDPVPEIRRDHFEEAMRFARRSVSDNDIRKY-EMFAQTLQQSRGFG 769
Query: 637 PFGFGQNNQ 645
F F NQ
Sbjct: 770 SFRFPSGNQ 778
Score = 163 bits (413), Expect = 1e-39
Identities = 100/285 (35%), Positives = 157/285 (55%), Gaps = 8/285 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V +DD+G + E+V LP+R P LF + +P +GILL+GPPGTGKTL+A+A+A
Sbjct: 201 VGYDDVGGCRKQLAQIKEMVELPLRHPALFKAIGV-KPPRGILLYGPPGTGKTLIARAVA 259
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E GA F ++G + SK G++E + F A K AP IIF+DE+D++ R
Sbjct: 260 NETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREKT-HG 318
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ ++ + DGL K+ ++++ ATNRP +D A+ R R R + + +PDA
Sbjct: 319 EVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIPDATG 376
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
R +IL+I K+ L D +++AN T G+ G+DL LC AA + +++ ++ D
Sbjct: 377 RLEILQIHTKNMKLADDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDLEDET 436
Query: 586 ITNVALRPL--NLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I + L +DDF + S+ PS + V W ++ G
Sbjct: 437 IDAEVMNSLAVTMDDFRWALSQSNPSALRETVVEVPQVTWEDIGG 481
>TERA_HUMAN (P55072) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
(Valosin-containing protein) (VCP)
Length = 805
Score = 186 bits (471), Expect = 2e-46
Identities = 117/309 (37%), Positives = 176/309 (56%), Gaps = 15/309 (4%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG LEDVK+ L ELV P+ P+ F + P KG+L +GPPG GKTL+AKA+A
Sbjct: 473 VTWEDIGGLEDVKRELQELVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 531
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E ANFIS+ G L + WFG++E + +F A + AP ++F DE+DS+ ARGG
Sbjct: 532 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 591
Query: 468 --EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
A R+ N+ + DG+ +K+N + I+GATNRP +D A++R RL + IY+ LPD
Sbjct: 592 GGGAADRVINQILTEMDGMSTKKN--VFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 649
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE-KAG 582
+R IL+ L+ + D + LA +T+G+SG+DL +C A ++E +E E +
Sbjct: 650 KSRVAILKANLRKSPVAKDVDLEFLAKMTNGFSGADLTEICQRACKLAIRESIESEIRRE 709
Query: 583 DNNITNVALRPLNLDDFV----QSKSKVGPSVAYDAVSVNELRKWNEMYGE--GGTRTKS 636
TN + + DD V + + A +VS N++RK+ EM+ + +R
Sbjct: 710 RERQTNPSAMEVEEDDPVPEIRRDHFEEAMRFARRSVSDNDIRKY-EMFAQTLQQSRGFG 768
Query: 637 PFGFGQNNQ 645
F F NQ
Sbjct: 769 SFRFPSGNQ 777
Score = 164 bits (414), Expect = 9e-40
Identities = 101/285 (35%), Positives = 157/285 (54%), Gaps = 8/285 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V +DDIG + E+V LP+R P LF + +P +GILL+GPPGTGKTL+A+A+A
Sbjct: 200 VGYDDIGGCRKQLAQIKEMVELPLRHPALFKAIGV-KPPRGILLYGPPGTGKTLIARAVA 258
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E GA F ++G + SK G++E + F A K AP IIF+DE+D++ R
Sbjct: 259 NETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREKT-HG 317
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ ++ + DGL K+ ++++ ATNRP +D A+ R R R + + +PDA
Sbjct: 318 EVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIPDATG 375
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
R +IL+I K+ L D +++AN T G+ G+DL LC AA + +++ ++ D
Sbjct: 376 RLEILQIHTKNMKLADDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDLEDET 435
Query: 586 ITNVALRPL--NLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I + L +DDF + S+ PS + V W ++ G
Sbjct: 436 IDAEVMNSLAVTMDDFRWALSQSNPSALRETVVEVPQVTWEDIGG 480
>SKD1_HUMAN (O75351) SKD1 protein (Vacuolar sorting protein 4b)
Length = 444
Score = 186 bits (471), Expect = 2e-46
Identities = 120/320 (37%), Positives = 178/320 (55%), Gaps = 42/320 (13%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
VK+ D+ LE K+AL E VILP++ P LFT P +GILLFGPPGTGK+ +AKA+A
Sbjct: 131 VKWSDVAGLEGAKEALKEAVILPIKFPHLFTGKRT--PWRGILLFGPPGTGKSYLAKAVA 188
Query: 408 TEAG-ANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFE 466
TEA + F S+S S L SKW G++EKL K LF A + P IIF+DE+DSL G+R E
Sbjct: 189 TEANNSTFFSISSSDLVSKWLGESEKLVKNLFQLARENKPSIIFIDEIDSLCGSR-SENE 247
Query: 467 HEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNR 526
EA RR++ EF+ G+ +N IL+LGATN P+ LD A+ RR +RIY+ LP+ R
Sbjct: 248 SEAARRIKTEFLVQMQGV-GVDNDGILVLGATNIPWVLDSAIRRRFEKRIYIPLPEPHAR 306
Query: 527 KKILRIIL-KHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLE-------- 577
+ ++ L +N + F +L TDGYSG+D+ + A +PV+++
Sbjct: 307 AAMFKLHLGTTQNSLTEADFRELGRKTDGYSGADISIIVRDALMQPVRKVQSATHFKKVR 366
Query: 578 -EEKAGDNNITNVALRP-------------------------LNLDDFVQSKSKVGPSVA 611
+A N++ + L P +++ D ++S S P+V
Sbjct: 367 GPSRADPNHLVDDLLTPCSPGDPGAIEMTWMDVPGDKLLEPVVSMSDMLRSLSNTKPTV- 425
Query: 612 YDAVSVNELRKWNEMYGEGG 631
+ + +L+K+ E +G+ G
Sbjct: 426 -NEHDLLKLKKFTEDFGQEG 444
>YB56_METJA (Q58556) Cell division cycle protein 48 homolog MJ1156
Length = 903
Score = 184 bits (468), Expect = 5e-46
Identities = 132/402 (32%), Positives = 213/402 (52%), Gaps = 27/402 (6%)
Query: 229 DRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLL 288
DR+IVI + +++L+ H + +D D + A+ G+ + C
Sbjct: 339 DREIVIGVPDREGRKEILQIHTRNMPLAEDVDLDYL------ADVTHGFVGADLAALCKE 392
Query: 289 PCVKGDKLCLPR---ESLEIAISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGE 345
++ + LP E+ EI L ++ ++LK++ + VP
Sbjct: 393 AAMRALRRVLPSIDLEAEEIPKEVLDNLKVTMDDFKEALKDVEPSAMREVLVE--VP--- 447
Query: 346 IGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKA 405
VK++DIG LE+VK+ L E V P++ E+F + RP KG+LLFGPPGTGKTL+AKA
Sbjct: 448 -NVKWEDIGGLEEVKQELREAVEWPLKAKEVFEKIGV-RPPKGVLLFGPPGTGKTLLAKA 505
Query: 406 LATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAF 465
+A E+GANFISV G + SKW G++EK + +F A + AP IIF DE+D++ RG
Sbjct: 506 VANESGANFISVKGPEIFSKWVGESEKAIREIFRKARQSAPCIIFFDEIDAIAPKRGRDL 565
Query: 466 EHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
T ++ N+ + DG+ +E + ++++ ATNRP +D A++R RL R I V +PD
Sbjct: 566 SSAVTDKVVNQLLTELDGM--EEPKDVVVIAATNRPDIIDPALLRPGRLDRVILVPVPDE 623
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGD 583
R I +I + NL D ++LA T+GY+G+D++ LC AA V+E + +
Sbjct: 624 KARLDIFKIHTRSMNLAEDVNLEELAKKTEGYTGADIEALCREAAMLAVRESIGKP---- 679
Query: 584 NNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNE 625
++ ++ L +++QS S + A + SV + K E
Sbjct: 680 ---WDIEVKLRELINYLQSISGTFRAAAVELNSVIKATKERE 718
Score = 183 bits (464), Expect = 1e-45
Identities = 110/285 (38%), Positives = 166/285 (57%), Gaps = 8/285 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG L++ K + E++ LPMR PELF + P KG+LL GPPGTGKTL+AKA+A
Sbjct: 176 VTYEDIGGLKEEVKKVREMIELPMRHPELFEKLGI-EPPKGVLLVGPPGTGKTLLAKAVA 234
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
EAGANF ++G + SK+ G+ E+ + +F A + AP IIF+DE+D++ R A
Sbjct: 235 NEAGANFYVINGPEIMSKYVGETEENLRKIFEEAEENAPSIIFIDEIDAIAPKRDEA-TG 293
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ + + DGL+ + +++++GATNRP LD A+ R R R I + +PD +
Sbjct: 294 EVERRLVAQLLTLMDGLKGR--GQVVVIGATNRPNALDPALRRPGRFDREIVIGVPDREG 351
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
RK+IL+I ++ L D D LA++T G+ G+DL LC AA R ++ +L
Sbjct: 352 RKEILQIHTRNMPLAEDVDLDYLADVTHGFVGADLAALCKEAAMRALRRVLPSIDLEAEE 411
Query: 586 ITNVALRPL--NLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I L L +DDF ++ V PS + + KW ++ G
Sbjct: 412 IPKEVLDNLKVTMDDFKEALKDVEPSAMREVLVEVPNVKWEDIGG 456
>TERA_PIG (P03974) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
(Valosin-containing protein) (VCP)
Length = 806
Score = 184 bits (468), Expect = 5e-46
Identities = 116/309 (37%), Positives = 176/309 (56%), Gaps = 15/309 (4%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V ++DIG LEDVK+ L +LV P+ P+ F + P KG+L +GPPG GKTL+AKA+A
Sbjct: 474 VTWEDIGGLEDVKRELQDLVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 532
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E ANFIS+ G L + WFG++E + +F A + AP ++F DE+DS+ ARGG
Sbjct: 533 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 592
Query: 468 --EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDA 523
A R+ N+ + DG+ +K+N + I+GATNRP +D A++R RL + IY+ LPD
Sbjct: 593 GGGAADRVINQILTEMDGMSTKKN--VFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 650
Query: 524 DNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE-KAG 582
+R IL+ L+ + D + LA +T+G+SG+DL +C A ++E +E E +
Sbjct: 651 KSRVAILKANLRKSPVAKDVDLEFLAKMTNGFSGADLTEICQRACKLAIRESIESEIRRE 710
Query: 583 DNNITNVALRPLNLDDFV----QSKSKVGPSVAYDAVSVNELRKWNEMYGE--GGTRTKS 636
TN + + DD V + + A +VS N++RK+ EM+ + +R
Sbjct: 711 RERQTNPSAMEVEEDDPVPEIRRDHFEEAMRFARRSVSDNDIRKY-EMFAQTLQQSRGFG 769
Query: 637 PFGFGQNNQ 645
F F NQ
Sbjct: 770 SFRFPSGNQ 778
Score = 164 bits (414), Expect = 9e-40
Identities = 101/285 (35%), Positives = 157/285 (54%), Gaps = 8/285 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V +DDIG + E+V LP+R P LF + +P +GILL+GPPGTGKTL+A+A+A
Sbjct: 201 VGYDDIGGCRKQLAQIKEMVELPLRHPALFKAIGV-KPPRGILLYGPPGTGKTLIARAVA 259
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E GA F ++G + SK G++E + F A K AP IIF+DE+D++ R
Sbjct: 260 NETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREKT-HG 318
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ ++ + DGL K+ ++++ ATNRP +D A+ R R R + + +PDA
Sbjct: 319 EVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIPDATG 376
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
R +IL+I K+ L D +++AN T G+ G+DL LC AA + +++ ++ D
Sbjct: 377 RLEILQIHTKNMKLADDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDLEDET 436
Query: 586 ITNVALRPL--NLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I + L +DDF + S+ PS + V W ++ G
Sbjct: 437 IDAEVMNSLAVTMDDFRWALSQSNPSALRETVVEVPQVTWEDIGG 481
>CC48_YEAST (P25694) Cell division control protein 48
Length = 835
Score = 183 bits (464), Expect = 1e-45
Identities = 97/239 (40%), Positives = 149/239 (61%), Gaps = 7/239 (2%)
Query: 346 IGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKA 405
+ V +DD+G L+++K+ L E V P+ P+ +T L P KG+L +GPPGTGKTL+AKA
Sbjct: 482 VNVTWDDVGGLDEIKEELKETVEYPVLHPDQYTKFGL-SPSKGVLFYGPPGTGKTLLAKA 540
Query: 406 LATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAF 465
+ATE ANFISV G L S W+G++E + +F A AP ++F+DE+DS+ ARGG+
Sbjct: 541 VATEVSANFISVKGPELLSMWYGESESNIRDIFDKARAAAPTVVFLDELDSIAKARGGSL 600
Query: 466 EHE--ATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLP 521
A+ R+ N+ + DG+ +K+N + ++GATNRP +D A++R RL + IYV LP
Sbjct: 601 GDAGGASDRVVNQLLTEMDGMNAKKN--VFVIGATNRPDQIDPAILRPGRLDQLIYVPLP 658
Query: 522 DADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK 580
D + R IL L+ L+P + +A T G+SG+DL + AA +++ +E +
Sbjct: 659 DENARLSILNAQLRKTPLEPGLELTAIAKATQGFSGADLLYIVQRAAKYAIKDSIEAHR 717
Score = 167 bits (424), Expect = 6e-41
Identities = 99/285 (34%), Positives = 160/285 (55%), Gaps = 8/285 (2%)
Query: 348 VKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALA 407
V +DDIG + E+V LP+R P+LF + +P +G+L++GPPGTGKTLMA+A+A
Sbjct: 211 VGYDDIGGCRKQMAQIREMVELPLRHPQLFKAIGI-KPPRGVLMYGPPGTGKTLMARAVA 269
Query: 408 TEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEH 467
E GA F ++G + SK G++E + F A K AP IIF+DE+DS+ R
Sbjct: 270 NETGAFFFLINGPEVMSKMAGESESNLRKAFEEAEKNAPAIIFIDEIDSIAPKRDKT-NG 328
Query: 468 EATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIR--RLPRRIYVDLPDADN 525
E RR+ ++ + DG++++ N ++++ ATNRP +D A+ R R R + + +PDA
Sbjct: 329 EVERRVVSQLLTLMDGMKARSN--VVVIAATNRPNSIDPALRRFGRFDREVDIGIPDATG 386
Query: 526 RKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNN 585
R ++LRI K+ L D + LA T GY G+D+ +LC AA + ++E ++ ++
Sbjct: 387 RLEVLRIHTKNMKLADDVDLEALAAETHGYVGADIASLCSEAAMQQIREKMDLIDLDEDE 446
Query: 586 ITNVALRPL--NLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYG 628
I L L +D+F + PS + V + W+++ G
Sbjct: 447 IDAEVLDSLGVTMDNFRFALGNSNPSALRETVVESVNVTWDDVGG 491
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 77,396,792
Number of Sequences: 164201
Number of extensions: 3382632
Number of successful extensions: 12417
Number of sequences better than 10.0: 829
Number of HSP's better than 10.0 without gapping: 556
Number of HSP's successfully gapped in prelim test: 274
Number of HSP's that attempted gapping in prelim test: 10982
Number of HSP's gapped (non-prelim): 1182
length of query: 645
length of database: 59,974,054
effective HSP length: 117
effective length of query: 528
effective length of database: 40,762,537
effective search space: 21522619536
effective search space used: 21522619536
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0135.9


