

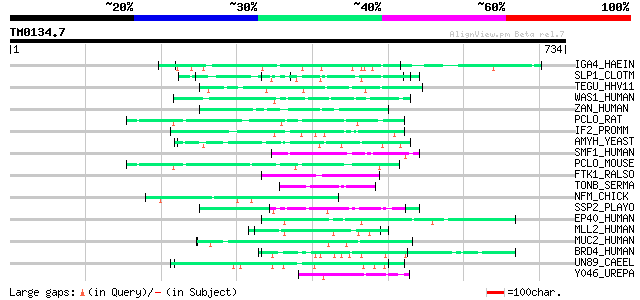
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.7
(734 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
IGA4_HAEIN (P45386) Immunoglobulin A1 protease precursor (EC 3.4... 66 3e-10
SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer... 64 1e-09
TEGU_HHV11 (P10220) Large tegument protein (Virion protein UL36) 62 5e-09
WAS1_HUMAN (Q92558) Wiskott-Aldrich syndrome protein family memb... 60 2e-08
ZAN_HUMAN (Q9Y493) Zonadhesin precursor 57 1e-07
PCLO_RAT (Q9JKS6) Piccolo protein (Multidomain presynaptic cytom... 57 2e-07
IF2_PROMM (Q7V5M4) Translation initiation factor IF-2 57 2e-07
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 57 2e-07
SMF1_HUMAN (O14497) SWI/SNF-related, matrix-associated, actin-de... 56 4e-07
PCLO_MOUSE (Q9QYX7) Piccolo protein (Presynaptic cytomatrix prot... 56 4e-07
FTK1_RALSO (Q8XRH0) DNA translocase ftsK 1 55 5e-07
TONB_SERMA (P26185) TonB protein 53 3e-06
NFM_CHICK (P16053) Neurofilament triplet M protein (160 kDa neur... 53 3e-06
SSP2_PLAYO (Q01443) Sporozoite surface protein 2 precursor 52 4e-06
EP40_HUMAN (Q96L91) E1A binding protein p400 (EC 3.6.1.-) (p400 ... 52 4e-06
MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia p... 52 6e-06
MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2) 51 1e-05
BRD4_HUMAN (O60885) Bromodomain-containing protein 4 (HUNK1 prot... 51 1e-05
UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89 (Uncoo... 50 2e-05
Y046_UREPA (Q9PR99) Hypothetical protein UU046 50 3e-05
>IGA4_HAEIN (P45386) Immunoglobulin A1 protease precursor (EC
3.4.21.72) (IGA1 protease)
Length = 1849
Score = 66.2 bits (160), Expect = 3e-10
Identities = 81/356 (22%), Positives = 137/356 (37%), Gaps = 48/356 (13%)
Query: 197 EDLSTIVSDVFTSTSLKKMGLVKK------KVAPAHEDTSDEKKKKKA------DKSKRK 244
ED T+ ++ T+ + + G ++ K P T E + +K DK +
Sbjct: 1133 EDQPTVEANTQTNEATQSEGKTEETQTAETKSEPTESVTVSENQPEKTVSQSTEDKVVVE 1192
Query: 245 HEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKC 304
EE K + AP QV E P + EPA + + T+A ++ Q+
Sbjct: 1193 KEEKAKVETEETQKAP-------QVTSKEPPKQAEPAPEEVPTDTNAEEAQALQQTQPTT 1245
Query: 305 VVTESNDDLNSLDALPISTLLNRT-ATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEA 363
V NS A +T A +TP++ A QPT + + + QE
Sbjct: 1246 VAAAETTSPNSKPAEETQQPSEKTNAEPVTPVVSENTATQPTETEETAKVE--KEKTQEV 1303
Query: 364 PLWNMLQTPRPSEPTSPITIQY---------NSLTSEPIILDQPEPEPRPSDHSVPRASE 414
P ++P+ +P + Q N LT++ + QP+ +P+ +VP E
Sbjct: 1304 PQVASQESPKQEQPAAKPQAQTKPQAEPARENVLTTKNVGEPQPQAQPQTQSTAVPTTGE 1363
Query: 415 RPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQ---QVQPDQVQQEPI----QPE 467
A P P+ + + +P ++ +P +Q V +Q +E QP
Sbjct: 1364 TAANSKPAAKPQAQAKPQTEPARE-NVSTVNTKEPQSQTSATVSTEQPAKETSSNVEQPA 1422
Query: 468 PETSVSNHSSV-------RSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSS 516
PE S++ S+ +S PQ+ET N +P T S SE++ S
Sbjct: 1423 PENSINTGSATTMTETAEKSDKPQMETVTEN--DRQPEANTVADNSVANNSESSES 1476
Score = 50.8 bits (120), Expect = 1e-05
Identities = 100/508 (19%), Positives = 186/508 (35%), Gaps = 71/508 (13%)
Query: 220 KKVA--PAHEDTSDEKKKKKADKSKRKHEESTKPDAPS------DGDAPPPKKKKKQVRI 271
+KVA P E+ + K +++A + ++ E K D P+ +A + K ++ +
Sbjct: 1100 EKVAENPPQENETVAKNEQEATEPTPQNGEVAKEDQPTVEANTQTNEATQSEGKTEETQT 1159
Query: 272 VEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQ 331
E T+ EP V + T S QS+ VV E + + T + A Q
Sbjct: 1160 AE--TKSEPTESVTVSENQPEKTVS--QSTEDKVVVEKEEKAK------VETEETQKAPQ 1209
Query: 332 LT---PILEFQPA-EQPTSPPNSPRSSFFQPSQQEAPLWNMLQTP--RPSEPTSPITIQY 385
+T P + +PA E+ + N+ + Q +Q +P +P+E T + +
Sbjct: 1210 VTSKEPPKQAEPAPEEVPTDTNAEEAQALQQTQPTTVAAAETTSPNSKPAEETQQPSEKT 1269
Query: 386 NSLTSEPIILDQPEPEPRPSDHSVP----RASERPALPSPRRYPRPRPERLVDPDHPIQA 441
N+ P++ + +P ++ + + E P + S + +P QA
Sbjct: 1270 NAEPVTPVVSENTATQPTETEETAKVEKEKTQEVPQVASQESPKQEQPAAKPQAQTKPQA 1329
Query: 442 DPLHEADPLAQQVQPDQVQQEPIQPE---PETSVSNHSSVRSPHPQVETSEPNLGTSEPN 498
+P E + V Q Q +P P T + +S + PQ + ++P + N
Sbjct: 1330 EPARENVLTTKNVGEPQPQAQPQTQSTAVPTTGETAANSKPAAKPQAQ-AKPQTEPAREN 1388
Query: 499 VQTFDVGSPQGASEANSSNHPTSPETNLSIVPYTYLRPNSLSECISVFNHEASLMIRNVQ 558
V T + PQ + A S + ET+ ++ E S+ +
Sbjct: 1389 VSTVNTKEPQSQTSATVSTEQPAKETSSNVEQPA---------------PENSINTGSAT 1433
Query: 559 GHTDLSENSDSVAEEWNRLSTWLVAQVPVLMSHLTAERDQRIEAARQRFNRRVAMHEQQQ 618
T+ +E SD E T + R A + VA + +
Sbjct: 1434 TMTETAEKSDKPQME-------------------TVTENDRQPEANTVADNSVANNSESS 1474
Query: 619 RTKLLEAVEEARRKKEQAEE---AARQEAAQAENARLEAERLEVEAEALRGQALAPVVFT 675
+K ++ K+ AEE A+ QE + R +++ + PV
Sbjct: 1475 ESKSRRRRSVSQPKETSAEETTVASTQETTVDNSVSTPKPRSRRTRRSVQTNSYEPVELP 1534
Query: 676 PVTSDSTQAPHSAQNVPSTSSQPSSSRL 703
+++ + S NV +SQP+ L
Sbjct: 1535 TENAENAENVQSGNNV--ANSQPALRNL 1560
Score = 43.5 bits (101), Expect = 0.002
Identities = 70/334 (20%), Positives = 124/334 (36%), Gaps = 46/334 (13%)
Query: 232 EKKKKKADKSKRKHEESTKPDAPS-----------DGDAPPPKKKKKQVRIVEKP-TRVE 279
EK+ + D + + DAPS + PPP + E+P TR
Sbjct: 999 EKRNQTVDTTNITTPNDIQADAPSAQSNNEEIARVETPVPPPAPATESAIASEQPETRPA 1058
Query: 280 PAAQVIRMKTS-ARVTRSAVQS---------SSKCVVTESNDDLNSLDALPISTLLN--R 327
AQ +T+ A T +A +S +S+ V +E+ + + T+ +
Sbjct: 1059 ETAQPAMEETNTANSTETAPKSDTATQTENPNSESVPSETTEKVAENPPQENETVAKNEQ 1118
Query: 328 TATQLTPIL-EFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYN 386
AT+ TP E +QPT N+ + + +Q E SEPT +T+ N
Sbjct: 1119 EATEPTPQNGEVAKEDQPTVEANTQTN---EATQSEGKTEETQTAETKSEPTESVTVSEN 1175
Query: 387 ------SLTSEPIILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQ 440
S ++E ++ + E + + +A + + P++ P PE + + +
Sbjct: 1176 QPEKTVSQSTEDKVVVEKEEKAKVETEETQKAPQVTSKEPPKQ-AEPAPEEVPTDTNAEE 1234
Query: 441 ADPLHEADPL----AQQVQPD-------QVQQEPIQPEPETSVSNHSSVRSPHPQVETSE 489
A L + P A+ P+ Q E EP T V + ++ P ET++
Sbjct: 1235 AQALQQTQPTTVAAAETTSPNSKPAEETQQPSEKTNAEPVTPVVSENTATQPTETEETAK 1294
Query: 490 PNLGTSEPNVQTFDVGSPQGASEANSSNHPTSPE 523
++ Q SP+ A T P+
Sbjct: 1295 VEKEKTQEVPQVASQESPKQEQPAAKPQAQTKPQ 1328
>SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer
layer protein B) (S-layer protein 1)
Length = 1664
Score = 63.9 bits (154), Expect = 1e-09
Identities = 71/278 (25%), Positives = 103/278 (36%), Gaps = 38/278 (13%)
Query: 246 EESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCV 305
+E T D P+ D P P ++PT E + I T + + + +
Sbjct: 786 DEPTPSDEPTPSDEPTPS---------DEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDE 836
Query: 306 VTESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPL 365
T S++ S + P + T PI P+++PT P + P PS + P
Sbjct: 837 PTPSDEPTPSDEPTP-------SETPEEPIPTDTPSDEPT-PSDEP-----TPSDEPTP- 882
Query: 366 WNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPEPRPSDHSVPRASERPALPSPRRYP 425
P PS+ +P + EPI D P EP PSD P S+ P PS P
Sbjct: 883 ---SDEPTPSDEPTP-----SETPEEPIPTDTPSDEPTPSDEPTP--SDEPT-PSDEPTP 931
Query: 426 RPRPERLVDPDHPIQAD-PLHEADPLAQQVQPDQ--VQQEPIQPEPETSVSNHSSVRSPH 482
P P+ PI D P E P + D+ EP + T + P
Sbjct: 932 SDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPT 991
Query: 483 PQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPT 520
P ET E + T P+ + P + E S+ PT
Sbjct: 992 PS-ETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPT 1028
Score = 62.0 bits (149), Expect = 5e-09
Identities = 75/316 (23%), Positives = 115/316 (35%), Gaps = 27/316 (8%)
Query: 224 PAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQ 283
P+ E T E ++ + +E T D P+ D P P + ++PT E +
Sbjct: 802 PSDEPTPSETPEEPIP-TDTPSDEPTPSDEPTPSDEPTPSDEPTPS---DEPTPSETPEE 857
Query: 284 VIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQ 343
I T + + + + T S++ S + P T T TP E P+++
Sbjct: 858 PIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTD-TPSDEPTPSDE 916
Query: 344 PT-----SPPNSPRSSFFQPSQQEAPLWNMLQTPRPS-EPT---SPITIQYNSLTSEPII 394
PT +P + P S +P+ E P + T PS EPT P + + EP
Sbjct: 917 PTPSDEPTPSDEPTPSD-EPTPSETPE-EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTP 974
Query: 395 LDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQV 454
D+P P P+ P SE P P P P P +P + P E P +
Sbjct: 975 SDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPT 1034
Query: 455 QPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEAN 514
D +P P + + P P S+ + EP P + E
Sbjct: 1035 PSD-------EPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPS----DEPTPSDEPT 1083
Query: 515 SSNHPTSPETNLSIVP 530
S+ PT ET +P
Sbjct: 1084 PSDEPTPSETPEEPIP 1099
Score = 60.8 bits (146), Expect = 1e-08
Identities = 75/327 (22%), Positives = 117/327 (34%), Gaps = 25/327 (7%)
Query: 224 PAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQ 283
P+ E T E ++ + +E T D P+ D P P + ++PT E +
Sbjct: 1127 PSDEPTPSETPEEPIP-TDTPSDEPTPSDEPTPSDEPTPSDEPTPS---DEPTPSETPEE 1182
Query: 284 VIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQ 343
I T + + + + T S++ S + P T T TP E P+++
Sbjct: 1183 PIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTD-TPSDEPTPSDE 1241
Query: 344 PT-----SPPNSPRSSFFQPSQQEAPLWNMLQTPRPS-EPT---SPITIQYNSLTSEPII 394
PT +P + P S +P+ E P + T PS EPT P + + EP
Sbjct: 1242 PTPSDEPTPSDEPTPSD-EPTPSETPE-EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTP 1299
Query: 395 LDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQV 454
D+P P P+ P SE P P P P P +P + P E P + +
Sbjct: 1300 SDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTP-SDEP 1358
Query: 455 QPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEAN 514
P +P P + + +P P T+ P + G G
Sbjct: 1359 TPSDEPTPSDEPTPSETPEEPTPTTTPTPTPSTT--------PTSGSGGSGGSGGGGGGG 1410
Query: 515 SSNHPTSPETNLSIVPYTYLRPNSLSE 541
PTSP + P + P + E
Sbjct: 1411 GGTVPTSPTPTPTSKPTSTPAPTEIEE 1437
Score = 59.7 bits (143), Expect = 3e-08
Identities = 71/312 (22%), Positives = 111/312 (34%), Gaps = 45/312 (14%)
Query: 224 PAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQ 283
P+ E T E ++ + +E T D P+ D P P ++PT +
Sbjct: 931 PSDEPTPSETPEEPIP-TDTPSDEPTPSDEPTPSDEPTPS---------DEPTPSDEPTP 980
Query: 284 VIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQ 343
S T S T S++ S + P + + TP E P+++
Sbjct: 981 SDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTP--------SDEPTPSDEPTPSDE 1032
Query: 344 PTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPEPR 403
PT P + P S +P+ E P + T PS+ +P + EP D+P P
Sbjct: 1033 PT-PSDEPTPSD-EPTPSETPE-EPIPTDTPSDEPTP--------SDEPTPSDEPTPSDE 1081
Query: 404 PSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEP 463
P+ P SE P P P P P +P + P E P + + P + +EP
Sbjct: 1082 PTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTP-SDEPTPSETPEEP 1140
Query: 464 I---------------QPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQ 508
I P E + S+ + ET E + T P+ + P
Sbjct: 1141 IPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPT 1200
Query: 509 GASEANSSNHPT 520
+ E S+ PT
Sbjct: 1201 PSDEPTPSDEPT 1212
Score = 56.6 bits (135), Expect = 2e-07
Identities = 54/198 (27%), Positives = 75/198 (37%), Gaps = 24/198 (12%)
Query: 334 PILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPI 393
PI P+++PT P + P PS + P P PS+ +P + EPI
Sbjct: 772 PIPTDTPSDEPT-PSDEP-----TPSDEPTP----SDEPTPSDEPTP-----SETPEEPI 816
Query: 394 ILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQAD-PLHEADPLAQ 452
D P EP PSD P S+ P PS P P P+ PI D P E P +
Sbjct: 817 PTDTPSDEPTPSDEPTP--SDEPT-PSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDE 873
Query: 453 QVQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASE 512
D EP + T + +P + T P+ P+ + P + E
Sbjct: 874 PTPSD----EPTPSDEPTPSDEPTPSETPEEPIPTDTPS-DEPTPSDEPTPSDEPTPSDE 928
Query: 513 ANSSNHPTSPETNLSIVP 530
S+ PT ET +P
Sbjct: 929 PTPSDEPTPSETPEEPIP 946
Score = 47.4 bits (111), Expect = 1e-04
Identities = 43/170 (25%), Positives = 62/170 (36%), Gaps = 22/170 (12%)
Query: 372 PRPSEPTSPITIQYNSLTSEPIILDQPEP--EPRPSDHSV----PRASERPALPSPRRYP 425
P P + S I ++ + EP D+P P EP PSD P SE P P P P
Sbjct: 762 PAPIKAASDEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTP 821
Query: 426 RPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEPI---------------QPEPET 470
P +P + P E P + + P + +EPI P E
Sbjct: 822 SDEPTPSDEPTPSDEPTPSDEPTP-SDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEP 880
Query: 471 SVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPT 520
+ S+ + ET E + T P+ + P + E S+ PT
Sbjct: 881 TPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPT 930
>TEGU_HHV11 (P10220) Large tegument protein (Virion protein UL36)
Length = 3164
Score = 62.0 bits (149), Expect = 5e-09
Identities = 78/318 (24%), Positives = 124/318 (38%), Gaps = 34/318 (10%)
Query: 251 PDAPSDGDAPP-------PKKKKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSK 303
P P D PP P + ++ + P P V R+TR AV S S
Sbjct: 2754 PALPIDPVPPPVATGTVLPGGENRRPPLTSGPAPTPPRVPV--GGPQRRLTRPAVASLS- 2810
Query: 304 CVVTESNDDLNSL--DALPISTLLNRTATQLTPILEF-----QPAEQPTSPP--NSPRSS 354
ES + L S A P + +L R + T PA QP +PP + P +
Sbjct: 2811 ----ESRESLPSPWDPADPTAPVLGRNPAEPTSSSPAGPSPPPPAVQPVAPPPTSGPPPT 2866
Query: 355 FFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNS---LTSEPIILDQPEPEPRPSDHSVPR 411
+ AP + + P +P + T LT + QP+P+P+P P+
Sbjct: 2867 YLTLEGGVAPGGPVSRRPTTRQPVATPTTSARPRGHLTVSRLSAPQPQPQPQPQPQPQPQ 2926
Query: 412 ASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEPIQPEPE-- 469
+P P P+ P+P+P+ P Q P + P Q QP Q QP+P+
Sbjct: 2927 PQPQPQ-PQPQPQPQPQPQPQPQPQPQPQPQPQPQPQP---QPQPQPQPQPQPQPQPQNG 2982
Query: 470 -TSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPTSPETNLSI 528
+ + +VR PQ S P S N +T S + ++ + H + +S+
Sbjct: 2983 HVAPGEYPAVRFRAPQNRPSVP-ASASSTNPRTGSSLSGVSSWASSLALHIDATPPPVSL 3041
Query: 529 VPYTYLRPNSLSECISVF 546
+ Y+ + S+ S+F
Sbjct: 3042 LQTLYVSDDEDSDATSLF 3059
>WAS1_HUMAN (Q92558) Wiskott-Aldrich syndrome protein family member
1 (WASP-family protein member 1) (Verprolin homology
domain-containing protein 1)
Length = 559
Score = 60.5 bits (145), Expect = 2e-08
Identities = 76/336 (22%), Positives = 123/336 (35%), Gaps = 46/336 (13%)
Query: 217 LVKKKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPT 276
L K+K+ ED EK+K+K R HE P AP D ++++ ++ + P
Sbjct: 160 LWKEKMLQDTEDKRKEKRKQKQKNLDRPHEPEKVPRAPHD-------RRREWQKLAQGPE 212
Query: 277 RVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPIL 336
E A ++ ++ + + D SL ALP S ++ T+ +
Sbjct: 213 LAEDDANLLHKHIEVANGPASHFETRPQTYVDHMDGSYSLSALPFSQ-MSELLTRAEERV 271
Query: 337 EFQPAEQPTSPP----------------------NSPRSSFFQPSQQEAPLWNMLQTPRP 374
+P E P PP N P+S P+ P++ + TP P
Sbjct: 272 LVRPHEPPPPPPMHGAGDAKPIPTCISSATGLIENRPQS----PATGRTPVF-VSPTPPP 326
Query: 375 SEPTSPITIQYNSLTSEPIILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVD 434
P P + +SL + + P P P A + PA+P P P ++
Sbjct: 327 PPPPLPSALSTSSLRAS--MTSTPPPPVPPPPPPPATALQAPAVPPPPA-PLQIAPGVLH 383
Query: 435 PDHPIQADPLHEADPLAQQVQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGT 494
P P A PL + P + P + P+ P P+ V + P P P +
Sbjct: 384 PAPPPIAPPLVQPSPPVARAAP-VCETVPVHPLPQGEV---QGLPPPPPPPPLPPPGIRP 439
Query: 495 SEPNVQTFDVGSPQGASEANSSNHPTSPETNLSIVP 530
S P T P G S T+P ++ ++P
Sbjct: 440 SSPVTVTALAHPPSGLHPTPS----TAPGPHVPLMP 471
>ZAN_HUMAN (Q9Y493) Zonadhesin precursor
Length = 2812
Score = 57.4 bits (137), Expect = 1e-07
Identities = 54/253 (21%), Positives = 99/253 (38%), Gaps = 25/253 (9%)
Query: 252 DAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESND 311
+ PS P K+K EKPT + K + + + S ++TE
Sbjct: 575 EKPSVTTEKPTVPKEKPTIPTEKPTISTEKPTIPSEKPNMPSEKPTIPSEKPTILTEKPT 634
Query: 312 DLNSLDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQT 371
+ +P T + P + P E+PT+P +S +P ++ T
Sbjct: 635 IPSEKPTIPSE---KPTISTEKPTV---PTEEPTTPTEETTTSMEEP---------VIPT 679
Query: 372 PRPSEPTSPITI--QYNSLTSEPIILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRP 429
+PS PT +I + +++ E I+ +P P ++P +E+P +P+ + P
Sbjct: 680 EKPSIPTEKPSIPTEKPTISMEETIISTEKPTISPEKPTIP--TEKPTIPTEKSTISPEK 737
Query: 430 ERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEP-IQPEPETSVSNHSSVRSPHPQVETS 488
P P + + P +P ++P I PE T + ++ + P + T
Sbjct: 738 -----PTTPTEKPTIPTEKPTISPEKPTTPTEKPTISPEKLTIPTEKPTIPTEKPTIPTE 792
Query: 489 EPNLGTSEPNVQT 501
+P + T EP T
Sbjct: 793 KPTISTEEPTTPT 805
Score = 31.6 bits (70), Expect = 7.8
Identities = 61/292 (20%), Positives = 99/292 (33%), Gaps = 46/292 (15%)
Query: 219 KKKVAPAHEDT-SDEKKKKKADKSKRKHEESTKP--DAPSDGDAPPPKKKKKQVRIVEKP 275
+K P + T EK +K EE T P + + P +K + E
Sbjct: 771 EKLTIPTEKPTIPTEKPTIPTEKPTISTEEPTTPTEETTISTEKPSIPMEKPTLPTEETT 830
Query: 276 TRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPI 335
T VE + K + + + + + + TE +P L T PI
Sbjct: 831 TSVEETT-ISTEKLTIPMEKPTISTEKPTIPTEKPTISPEKLTIPTEKLTIPTEKPTIPI 889
Query: 336 LEFQ--------PAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITI--QY 385
E P E+PT P P S T +P+ PT TI +
Sbjct: 890 EETTISTEKLTIPTEKPTISPEKPTIS----------------TEKPTIPTEKPTIPTEE 933
Query: 386 NSLTSEPIILDQPEPEPRPSDHSVPR-----ASERPALPS-----PRRYPRPRPERLVDP 435
++++E + + +P P ++P ++E+P +P+ P P E+ P
Sbjct: 934 TTISTEKLTIPTEKPTISPEKLTIPTEKPTISTEKPTIPTEKLTIPTEKPTIPTEKPTIP 993
Query: 436 DHPIQA----DPLHEADPLAQQVQPDQVQQEPIQPE--PETSVSNHSSVRSP 481
+ A P A LA V P+ T+ S S+ R P
Sbjct: 994 TEKLTALRPPHPSPTATGLAALVMSPHAPSTPMTSVILGTTTTSRSSTERCP 1045
>PCLO_RAT (Q9JKS6) Piccolo protein (Multidomain presynaptic
cytomatrix protein)
Length = 5085
Score = 57.0 bits (136), Expect = 2e-07
Identities = 93/400 (23%), Positives = 149/400 (37%), Gaps = 57/400 (14%)
Query: 155 IRKSRTTASIKSAIKYIPFGRLLSDLFI-ESKFIEDLIKAGCTEDLSTIVSDVFTSTSLK 213
+ KSRTT + +S K +P GR S + + ESK D + + + SDV +++
Sbjct: 106 LSKSRTTDTFRSEQK-LP-GRSPSTISLKESKSRTDFKEEYKSSMMPGFFSDVNPLSAVS 163
Query: 214 KM-------GLVKKKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKK 266
+ L+ A E T KK+K K + K E KP P PK++
Sbjct: 164 SVVNKFNPFDLISDSEASQEETT---KKQKVVQKEQGKSEGMAKPPLQQPSPKPIPKQQG 220
Query: 267 KQVRIVEKPTRVEPAA--QVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTL 324
+ ++++ + + + Q ++K A T Q S + A P +
Sbjct: 221 QVKEVIQQDSSPKSVSSQQAEKVKPQAPGTGKPSQQSPAQTPAQ--------QASPGKPV 272
Query: 325 LNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQP---------------SQQEAPLWNML 369
+ + + + PA+ P P + +S P SQQ P ++
Sbjct: 273 AQQPGSAKATVQQPGPAKSPAQPAGTGKSPAQPPAKTPGQQAGLEKTSSSQQPGPK-SLA 331
Query: 370 QTPRPSE-PTSPITIQYNSLTSEPIILDQPEPEPRPSDHS-VPRASERPALPS-PRRYPR 426
QTP + P P+ S +P P +P P + VP ++ PA S P + P
Sbjct: 332 QTPGHGKFPLGPV----KSPAQQPGTAKHPAQQPGPQTAAKVPGPTKTPAQQSGPGKTPA 387
Query: 427 PRP-ERLVDPDHPIQADPLHEADPLAQQVQPDQ---VQQEPIQPEPETSVSNHSSVRSPH 482
+P P PI A P P+A + QP Q + +P QP P + P
Sbjct: 388 QQPGPTKPSPQQPIPAKP-QPQQPVATKTQPQQSAPAKPQPQQPAPAKPQPQQPTPAKPQ 446
Query: 483 PQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPTSP 522
PQ T ++P Q PQ + H P
Sbjct: 447 PQPPT------PAKPQPQPPTATKPQPQPPTATKPHHQQP 480
Score = 43.1 bits (100), Expect = 0.003
Identities = 66/306 (21%), Positives = 109/306 (35%), Gaps = 38/306 (12%)
Query: 220 KKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDA------PPPKKKKKQVRIVE 273
+KV P T ++ A ++ + KP A G A P P K Q
Sbjct: 241 EKVKPQAPGTGKPSQQSPA-QTPAQQASPGKPVAQQPGSAKATVQQPGPAKSPAQPAGTG 299
Query: 274 KPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLT 333
K + +P A+ + T S+ Q K + L P+ + + T
Sbjct: 300 K-SPAQPPAKTPGQQAGLEKTSSSQQPGPKSLAQTPGHGKFPLG--PVKSPAQQPGTAKH 356
Query: 334 PILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPI 393
P QP Q + P + P+QQ P Q P P++P+ +PI
Sbjct: 357 PAQ--QPGPQTAAKVPGPTKT---PAQQSGPGKTPAQQPGPTKPS----------PQQPI 401
Query: 394 ILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQ 453
+P+P+ + + P+ S PA P P++ +P+ P P A P P +
Sbjct: 402 PA-KPQPQQPVATKTQPQQS-APAKPQPQQPAPAKPQ----PQQPTPAKP-QPQPPTPAK 454
Query: 454 VQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEA 513
QP QP+P T+ H H Q ++P+ ++ G P
Sbjct: 455 PQPQPPTATKPQPQPPTATKPH------HQQPGLAKPSAQQPTKSISQTVTGRPLQPPPT 508
Query: 514 NSSNHP 519
+++ P
Sbjct: 509 SAAQTP 514
Score = 42.0 bits (97), Expect = 0.006
Identities = 40/162 (24%), Positives = 64/162 (38%), Gaps = 25/162 (15%)
Query: 348 PNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPEPRPSDH 407
P+SP+ + P AP ++P PS+ SP + EP PS
Sbjct: 589 PSSPQPT---PKAATAPTATASKSPVPSQQASP------------------KKEP-PSKQ 626
Query: 408 SVPRASERPALPSPRRYPRP-RPERLVDPDHPIQADPLHEADP-LAQQVQPDQVQQEPIQ 465
P+A E P P++ P P +P P ++ LH P A Q+ + EP
Sbjct: 627 DSPKALESKKPPEPKKPPEPKKPPEPKKPPPLVKQPTLHGPTPATAPQLPVAEALPEPAP 686
Query: 466 PEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSP 507
P+ E S ++P VE +P + + ++Q+ P
Sbjct: 687 PK-EPSGPLPEQAKAPVGDVEPKQPKMTETRADIQSSSTTKP 727
Score = 40.4 bits (93), Expect = 0.017
Identities = 72/331 (21%), Positives = 114/331 (33%), Gaps = 86/331 (25%)
Query: 223 APAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAP-PPKKKKKQVRIVEKPTRVEPA 281
+P + KK+ + + K ES KP P P P + KK +V++PT P
Sbjct: 609 SPVPSQQASPKKEPPSKQDSPKALESKKPPEPKKPPEPKKPPEPKKPPPLVKQPTLHGPT 668
Query: 282 AQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQPA 341
TA QL P+ E P
Sbjct: 669 P--------------------------------------------ATAPQL-PVAEALPE 683
Query: 342 EQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPE 401
P P+ P P Q +AP+ + ++ +P + IQ +S T I+ Q + +
Sbjct: 684 PAPPKEPSGP-----LPEQAKAPVGD-VEPKQPKMTETRADIQSSSTTKPDILSSQVQSQ 737
Query: 402 -------PRPSD-----HSVPRASERPALPSPRRYPRPRPE-RLVDPDHP--IQADPLHE 446
P +D S P E+ + PRP + +++ P DP H
Sbjct: 738 AQVKTASPLKTDSAKPSQSFPPTGEKTTPLDSKAMPRPASDSKIISQPGPGSESKDPKH- 796
Query: 447 ADPLAQQVQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGS 506
DP+ ++ +P + Q + P+PET V P + P G + P Q
Sbjct: 797 IDPIQKKDEPKKAQPKG-SPKPET-----KPVPKGSPTPSGTRPTAGQAAPPSQ--QPPK 848
Query: 507 PQGASEANSSN----------HPTSPETNLS 527
PQ S S N PT+P+ ++
Sbjct: 849 PQEQSRRFSLNLGGITDAPKSQPTTPQETVT 879
Score = 38.1 bits (87), Expect = 0.084
Identities = 107/598 (17%), Positives = 202/598 (32%), Gaps = 76/598 (12%)
Query: 151 LKECIRKSRTTASIKSAIKYIPFGRLLSDLFIESKFIEDLIKAGCTEDLSTIVSDVFTST 210
+KE + + ++ K +Y G ++D + + ++ ++ +G D + S
Sbjct: 3349 VKEEKQPKKRSSGAKVRGQYDEMGESVAD---DPRNLKKIVDSGVQTDDEETADRSYASR 3405
Query: 211 SLKKMGLVKKKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQV- 269
+ V V D D+ + +S+RK D+ ++GD P K V
Sbjct: 3406 RRRTKKSVDTSV---QTDDEDQDEWDMPSRSRRKARTGKYGDSTAEGDKTKPLSKVSSVA 3462
Query: 270 --RIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNR 327
+ E + EP + ARV + V+ E SL
Sbjct: 3463 VQTVAEISVQTEPVGTIRTPSIRARVD-AKVEIIKHISAPEKTYKGGSLGC--------- 3512
Query: 328 TATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNS 387
T+ + P TSPP + RP +P+ I Y+S
Sbjct: 3513 -QTETDSDTQSPPYLGATSPPKDKK--------------------RP----TPLEIGYSS 3547
Query: 388 --LTSEPIILDQPEPEPRPSDHSVPRASERPA---LPSPRRYPRPRPERLVDPDHPIQAD 442
L ++P + P P P P + P P+ Y +P P+ + P + D
Sbjct: 3548 SHLRADPTVQLAPSPPKSPKVLYSPISPLSPGNALEPAFVPYEKPLPDD-ISPQKVLHPD 3606
Query: 443 PLHEADPLAQQVQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTF 502
P A +Q+ P+P + ++ SS R+P E G + QT
Sbjct: 3607 MAKV--PPASPKTAKMMQRSMSDPKPLSPTADESS-RAPFQYSE------GFTTKGSQTM 3657
Query: 503 DVGSPQGASEANSSNHPTSPETNLSIVPYTYLRPNSLSECISVFNHEASLMIRNVQGHTD 562
Q + N P + + Y+ + S + +++++ D
Sbjct: 3658 TASGTQKKVKRTLPNPPPEEVSTGTQSTYSTMGTASRRRMCRTNTMARAKILQDIDRELD 3717
Query: 563 LSENSDSVAEEWNRLSTWLVAQVPVLMSHLTAERDQRIEAARQRFNRR--VAMHEQQQRT 620
L E + + + + E D ++ NRR + E+++R
Sbjct: 3718 LVERESA-----------KLRKKQAELDEEEKEIDAKLRYLEMGINRRKEALLKEREKRE 3766
Query: 621 K-LLEAVEEARRKKEQAEEAARQEAAQAENARLEAERLEVEAE---ALRGQALAPVVFTP 676
+ L+ V E R +E ++ + + +E R + E + Q P
Sbjct: 3767 RAYLQGVAEDRDYMSDSEVSSTRPSRVESQHGVERPRTAPQTEFSQFIPPQTQTEAQLVP 3826
Query: 677 VTSDSTQAPHSAQNVPSTSSQPSSSRLDIMEQRLNSHETLLLEMKQVLQELLRRSDKP 734
TS TQ +S+ +P+ + P + + +Q L + + + Q + S P
Sbjct: 3827 PTSPYTQYQYSSPALPTQAPTPYTQQSHFQQQTLYHQQVSPYQTQPTFQAVATMSFTP 3884
Score = 36.6 bits (83), Expect = 0.24
Identities = 49/243 (20%), Positives = 91/243 (37%), Gaps = 41/243 (16%)
Query: 220 KKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRVE 279
KK P+ +D+ + KK + K+ E P P + PPP K+ + T +
Sbjct: 619 KKEPPSKQDSPKALESKKPPEPKKPPE----PKKPPEPKKPPPLVKQPTLHGPTPATAPQ 674
Query: 280 -PAAQVIRMKTSARVTRSAVQSSSKCVV----------TESNDDLNSLDALPISTLLNRT 328
P A+ + + + +K V TE+ D+ S L ++
Sbjct: 675 LPVAEALPEPAPPKEPSGPLPEQAKAPVGDVEPKQPKMTETRADIQSSSTTKPDILSSQV 734
Query: 329 ATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQY-NS 387
+Q + P + ++ P S F P+ ++ + PRP+ + I+ S
Sbjct: 735 QSQ-AQVKTASPLKTDSAKP----SQSFPPTGEKTTPLDSKAMPRPASDSKIISQPGPGS 789
Query: 388 LTSEPIILD-----------QPEPEPRPSDHSVPRASERP---------ALPSPRRYPRP 427
+ +P +D QP+ P+P VP+ S P A P ++ P+P
Sbjct: 790 ESKDPKHIDPIQKKDEPKKAQPKGSPKPETKPVPKGSPTPSGTRPTAGQAAPPSQQPPKP 849
Query: 428 RPE 430
+ +
Sbjct: 850 QEQ 852
>IF2_PROMM (Q7V5M4) Translation initiation factor IF-2
Length = 1125
Score = 56.6 bits (135), Expect = 2e-07
Identities = 87/349 (24%), Positives = 132/349 (36%), Gaps = 59/349 (16%)
Query: 213 KKMGLVKKKVAPAHEDTSDEKKKKKADKS--KRKHEESTKPDAPSDGDAPPPKKK-KKQV 269
K +GL K V A E S K + S + K + APP K + +K +
Sbjct: 14 KDLGLENKDVLHAAEKLSIAAKSHSSSISDDEAKRIRGLLRQGSAANSAPPSKSEPEKTI 73
Query: 270 RIVEK--PTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNR 327
V+K PT ++ A IR TS SS+ + + N + P
Sbjct: 74 LSVKKAAPTAIKDVAPPIRKATS----------SSEISHVKPSAPANPIPTSPARPSTES 123
Query: 328 TATQLTPILEFQPAEQPTSPP--------NSPRSSFFQPSQQEAPLWNMLQTPRPS-EPT 378
A P P PTS P N+P S PS+ AP TPR + +P+
Sbjct: 124 VAHPAPPTRPANPTPTPTSSPPKTAARPINAPISRPAIPSRPTAP------TPRSANKPS 177
Query: 379 SPITIQ---------YNSLTSEPIILDQPEPEP----RPSDHSVPRASE---RPALPSPR 422
SP+ S TS+ + P P RP + P S +P++PS R
Sbjct: 178 SPVPPSAGGKDPRAGQTSTTSKATTVSGGGPRPKIVSRPQSPTAPGRSAPPAKPSIPSER 237
Query: 423 RYPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEPIQPEPETSVSNHSSV-RSP 481
+ P+P P P+ A P DP Q +PD+ ++ I P P + ++ + P
Sbjct: 238 KAPKPELVGRPKPKRPVVAPP-SRPDPEGQ--RPDK-KRPGISPRPIGGPNQRANTSQRP 293
Query: 482 HPQVETSEPNLGTSEPNVQTFD-VGSP-------QGASEANSSNHPTSP 522
+ + G T + VG P G++ +S+N P +P
Sbjct: 294 GAPIRQGKTRPGQPRSAGNTLELVGKPIRRDRSDAGSAGRDSNNRPGAP 342
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 56.6 bits (135), Expect = 2e-07
Identities = 77/324 (23%), Positives = 120/324 (36%), Gaps = 37/324 (11%)
Query: 219 KKKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRV 278
K+K P H DT+ KKK SK +++T P P P + PT
Sbjct: 282 KEKPTPPHHDTT-PCTKKKTTTSKTCTKKTTTP-------VPTPSSSTTESSSAPVPTPS 333
Query: 279 EPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEF 338
+ +SA VT S +SSS V T S+ S A S+ ++ +T
Sbjct: 334 SSTTE----SSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTS---- 385
Query: 339 QPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTS-PITIQYNSLTSEPIILDQ 397
E ++P +P SS + S AP+ T +E +S P+T +S P+
Sbjct: 386 STTESSSAPVPTPSSSTTESS--SAPV-----TSSTTESSSAPVTSSTTESSSAPVTSST 438
Query: 398 PEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDH--------PIQADPLHEADP 449
E P S +S P +P+P V P + E+
Sbjct: 439 TESSSAPVTSSTTESSSAP-VPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSS 497
Query: 450 LAQQVQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPN---LGTSEPNVQTFDVGS 506
+ P+ P P +S + SS +P P T+E + + +S + V +
Sbjct: 498 APVTSSTTESSSAPV-PTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPT 556
Query: 507 PQGASEANSSNHPTSPETNLSIVP 530
P ++ +SS TS T S P
Sbjct: 557 PSSSTTESSSTPVTSSTTESSSAP 580
Score = 46.2 bits (108), Expect = 3e-04
Identities = 68/319 (21%), Positives = 104/319 (32%), Gaps = 31/319 (9%)
Query: 223 APAHEDTSDEKKKKKADKSKRKHEESTKPDAPS---DGDAPPPKKKKKQVRIVEKPTRVE 279
AP T++ S E S+ P S AP P P
Sbjct: 540 APVTSSTTESSSAPVPTPSSSTTESSSTPVTSSTTESSSAPVPTPSSSTTESSSAPVPTP 599
Query: 280 PAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQ 339
++ A S+ SS VT S + +S S+ +++ P
Sbjct: 600 SSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSS 659
Query: 340 PAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTS-PITIQYNSLTSEPIILDQP 398
E ++P +P SS + S AP+ T +E +S P+T +S P+ P
Sbjct: 660 TTESSSAPVPTPSSSTTESSS--APV-----TSSTTESSSAPVTSSTTESSSAPV----P 708
Query: 399 EPEPRPSDHS---VPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEAD-PLAQQV 454
P ++ S VP S S P P P+ + + P+
Sbjct: 709 TPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPS 768
Query: 455 QPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEAN 514
P P +S + SS P P T+E ++ V +P +S
Sbjct: 769 SSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSVA---------PVPTPSSSSNIT 819
Query: 515 S---SNHPTSPETNLSIVP 530
S S+ P S T S VP
Sbjct: 820 SSAPSSTPFSSSTESSSVP 838
Score = 37.0 bits (84), Expect = 0.19
Identities = 56/263 (21%), Positives = 92/263 (34%), Gaps = 37/263 (14%)
Query: 289 TSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQPTSPP 348
TS T ++ S S + +++ + + S+ + T TP E+PT PP
Sbjct: 217 TSESSTTTSSTSESSTTTSSTSESSTTTSSTSESSTSSSTTAPATPTTTSCTKEKPT-PP 275
Query: 349 NSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPEPRPSDHS 408
+ + +P TP P T+P T + + TS+ P P PS S
Sbjct: 276 TTTSCTKEKP------------TP-PHHDTTPCT-KKKTTTSKTCTKKTTTPVPTPSS-S 320
Query: 409 VPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEPIQPEP 468
+S P P P P+ + + P+ P P
Sbjct: 321 TTESSSAPV-------PTPSSSTTESSSAPVTSSTTESS-------------SAPV-PTP 359
Query: 469 ETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPTSPETNLSI 528
+S + SS E+S + +S + V +P ++ +SS TS T S
Sbjct: 360 SSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSS 419
Query: 529 VPYTYLRPNSLSECISVFNHEAS 551
P T S S ++ E+S
Sbjct: 420 APVTSSTTESSSAPVTSSTTESS 442
>SMF1_HUMAN (O14497) SWI/SNF-related, matrix-associated,
actin-dependent regulator of chromatin subfamily F
member 1 (SWI-SNF complex protein p270) (B120)
Length = 1902
Score = 55.8 bits (133), Expect = 4e-07
Identities = 53/203 (26%), Positives = 88/203 (43%), Gaps = 21/203 (10%)
Query: 347 PPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLT--SEPIILDQPEPEPRP 404
PP + Q + P +N Q+P P + P + Q S T ++P QP+ +P
Sbjct: 69 PPYGQQGPSGYGQQGQTPYYNQ-QSPHPQQQQPPYSQQPPSQTPHAQPSYQQQPQSQPPQ 127
Query: 405 SDHSVPRASERPALPSPRRYPRPRP-ERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEP 463
S P S++P+ P ++ P P P ++ HP P + Q Q QQ+P
Sbjct: 128 LQSSQPPYSQQPSQPPHQQSPAPYPSQQSTTQQHPQSQPPYSQ-----PQAQSPYQQQQP 182
Query: 464 IQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPTS-- 521
QP P T +S ++ P PQ + S+ + + ++ S+A+S+ TS
Sbjct: 183 QQPAPST-LSQQAAY--PQPQSQQSQQTAYSQQRFPPPQELSQDSFGSQASSAPSMTSSK 239
Query: 522 ---PETNLSIVPYTYLRPNSLSE 541
+ NLS+ RP+SL +
Sbjct: 240 GGQEDMNLSL----QSRPSSLPD 258
Score = 43.1 bits (100), Expect = 0.003
Identities = 50/218 (22%), Positives = 75/218 (33%), Gaps = 50/218 (22%)
Query: 341 AEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEP 400
++QP S + S+ Q Q + P Q P +P+ P Q P
Sbjct: 103 SQQPPSQTPHAQPSYQQQPQSQPPQLQSSQPPYSQQPSQP--------------PHQQSP 148
Query: 401 EPRPSDHSVPRASERPALPSPRRYPRPR-PERLVDPDHPIQADPLHEA---DPLAQQVQP 456
P PS S + P P P+ + P + P P + +A P +QQ Q
Sbjct: 149 APYPSQQST--TQQHPQSQPPYSQPQAQSPYQQQQPQQPAPSTLSQQAAYPQPQSQQSQQ 206
Query: 457 DQVQQEPIQPEPETSVSNHSSVRSPHPQVETSE-----------------PNL------- 492
Q+ P E S + S S P + +S+ P+L
Sbjct: 207 TAYSQQRFPPPQELSQDSFGSQASSAPSMTSSKGGQEDMNLSLQSRPSSLPDLSGSIDDL 266
Query: 493 -----GTSEPNVQTFDVGSPQGASEANSSNHPTSPETN 525
G P V T + S QG ++N + P SP T+
Sbjct: 267 PMGTEGALSPGVSTSGISSSQG-EQSNPAQSPFSPHTS 303
>PCLO_MOUSE (Q9QYX7) Piccolo protein (Presynaptic cytomatrix
protein) (Aczonin) (Brain-derived HLMN protein)
Length = 5038
Score = 55.8 bits (133), Expect = 4e-07
Identities = 95/388 (24%), Positives = 142/388 (36%), Gaps = 49/388 (12%)
Query: 155 IRKSRTTASIKSAIKYIPFGRLLSDLFI-ESKFIEDLIKAGCTEDLSTIVSDVFTSTSLK 213
+ KSRTT + +S K +P GR S + + ESK D + + + S+V +++
Sbjct: 107 LSKSRTTDTFRSEQK-LP-GRSPSTISLKESKSRTDFKEEYKSSMMPGFFSEVNPLSAVS 164
Query: 214 KM-------GLVKKKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDA--PSDGDAPPPKK 264
+ L+ A E T KK+K A K + K E TKP PS P +
Sbjct: 165 SVVNKFNPFDLISDSEAVQEETT---KKQKVAQKDQGKSEGITKPSLQQPSPKLIPKQQG 221
Query: 265 KKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTL 324
K+V + P++ + Q + K A T Q S + + P
Sbjct: 222 PGKEVIPQDIPSKSVSSQQAEKTKPQAPGTAKPSQQSPAQTPAQQAKPVAQQPG-PAKAT 280
Query: 325 LNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSE-------- 376
+ + +P + P PP + + P+QQ LQ P P
Sbjct: 281 VQQPGPAKSPAQPAGTGKSPAQPPVTAKP----PAQQAGLEKTSLQQPGPKSLAQTPGQG 336
Query: 377 --PTSPITIQYNSLTSEPIILDQPEPEPRPSDHS-VPRASERPA-LPSPRRYPRPRP-ER 431
P P S +P P +P P S VP ++ PA L P + P +P
Sbjct: 337 KVPPGPA----KSPAQQPGTAKLPAQQPGPQTASKVPGPTKTPAQLSGPGKTPAQQPGPT 392
Query: 432 LVDPDHPIQADPLHEADPLAQQVQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVET---S 488
P PI A P Q QP + +P QP P H + P PQ T
Sbjct: 393 KPSPQQPIPAKP--------QPQQPVATKPQPQQPAPAKPQPQHPTPAKPQPQQPTPAKP 444
Query: 489 EPNLGT-SEPNVQTFDVGSPQGASEANS 515
+P T ++P Q +G P + S
Sbjct: 445 QPQQPTPAKPQPQHPGLGKPSAQQPSKS 472
Score = 43.9 bits (102), Expect = 0.002
Identities = 104/598 (17%), Positives = 206/598 (34%), Gaps = 78/598 (13%)
Query: 151 LKECIRKSRTTASIKSAIKYIPFGRLLSDLFIESKFIEDLIKAGCTEDLSTIVSDVFTST 210
+KE + + ++ K +Y G ++D + + ++ ++ +G D + S
Sbjct: 3303 VKEEKQPKKRSSGAKVRGQYDEMGESMAD---DPRNLKKIVDSGVQTDDEETADRTYASR 3359
Query: 211 SLKKMGLVKKKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVR 270
+ V V D D+ + +S+RK D+ ++GD P K
Sbjct: 3360 RRRTKKSVDTSV---QTDDEDQDEWDMPSRSRRKARTGKYGDSTAEGDKTKPPSK----- 3411
Query: 271 IVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTAT 330
V+ AVQ+ V E + L + ++ R
Sbjct: 3412 ----------------------VSSVAVQT-----VAEISVQTEPLGTIRTPSIRARVDA 3444
Query: 331 QLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNS--L 388
++ I E+ + + P Q P P+ + +P+ I Y+S L
Sbjct: 3445 KVEIIKHISAPEKTYKGGSLGCQTETDPDTQSPPYMGATSPPKDKKRPTPLEIGYSSSHL 3504
Query: 389 TSEPIILDQPEPEPRPSDHSVPRASERPA---LPSPRRYPRPRPERLVDPD---HPIQAD 442
++P + P P P P + P P+ Y +P P+ + P HP A
Sbjct: 3505 RADPTVQLAPSPPKSPKVLYSPISPLSPGHALEPAFVPYEKPLPDD-ISPQKVLHPDMAK 3563
Query: 443 PLHEADPLAQQVQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTF 502
+ A+ +Q +P+ P + S S S + S+ GT + +T
Sbjct: 3564 VPPASPKTAKMMQRSMSDPKPLSPTADES-SRAPFQYSEGFTAKGSQTTSGTQKKVKRTL 3622
Query: 503 DVGSPQGASEANSSNHPTSPETNLSIVPYTYLRPNSLSECISVFNHEASLMIRNVQGHTD 562
P+ AS S + T + R N+++ + +++++ D
Sbjct: 3623 PNPPPEEASTGTQSTYSTMGTASRR----RMCRTNTMAR---------AKILQDIDRELD 3669
Query: 563 LSENSDSVAEEWNRLSTWLVAQVPVLMSHLTAERDQRIEAARQRFNRR--VAMHEQQQRT 620
L E + + + + E D ++ NRR + E+++R
Sbjct: 3670 LVERESA-----------KLRKKQAELDEEEKEIDAKLRYLEMGINRRKEALLKEREKRE 3718
Query: 621 K-LLEAVEEARRKKEQAEEAARQEAAQAENARLEAERLEVEAE---ALRGQALAPVVFTP 676
+ L+ V E R +E ++ + + +E R + E + Q P
Sbjct: 3719 RAYLQGVAEDRDYMSDSEVSSTRPSRVESQHGIERPRTAPQTEFSQFIPPQTQTEAQLVP 3778
Query: 677 VTSDSTQAPHSAQNVPSTSSQPSSSRLDIMEQRLNSHETLLLEMKQVLQELLRRSDKP 734
TS TQ +S+ +P+ + P + + +Q L + + + Q + S P
Sbjct: 3779 PTSPYTQYQYSSPALPTQAPTPYTQQSHFQQQTLYHQQVSPYQTQPTFQAVATMSFTP 3836
Score = 43.9 bits (102), Expect = 0.002
Identities = 63/321 (19%), Positives = 121/321 (37%), Gaps = 47/321 (14%)
Query: 246 EESTKPDAPSDGDAPPPKKKKKQVRIVEKPT---------RVEPAAQVIRMKTSARVTRS 296
+ S K + PS D+P + KK +V++PT P A+ + + +
Sbjct: 593 QASPKKELPSKQDSPKAPESKKPPPLVKQPTLHGPTPATAPQPPVAEALPKPAPPKKPSA 652
Query: 297 AVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQ---PAEQPTSPP----N 349
A+ +K V + + + AT IL Q A+ T+PP +
Sbjct: 653 ALPEQAKAPVADVEPKQPKTTETLTDSPSSAAATSKPAILSSQVQAQAQVTTAPPLKTDS 712
Query: 350 SPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPE-------P 402
+ S F P+ + PRP+ + ++ + S+ + + EP+ P
Sbjct: 713 AKTSQSFPPTGDTITPLDSKAMPRPASDSKIVSHPGPTSESKDPVQKKEEPKKAQTKVTP 772
Query: 403 RPSDHSVPRASERP---------ALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQ 453
+P VP+ S P A P ++ P+P PE+ + + +A P +Q
Sbjct: 773 KPDTKPVPKGSPTPSGTRPTTGQATPQSQQPPKP-PEQ--SRRFSLNLGGIADA-PKSQP 828
Query: 454 VQPDQVQQEPIQPEPETSVSNHSSV------RSPHPQVETSEPNLGTSEPNVQTFDVGSP 507
P + + + S S++ ++PHPQ + P+ P+ G P
Sbjct: 829 TTPQETVTGKLFGFGASIFSQASNLISTAGQQAPHPQTGPAAPSKQAPPPSQTLAAQGPP 888
Query: 508 QGASEANSSNHPTSPETNLSI 528
+ ++ HP++P ++
Sbjct: 889 K-----STGQHPSAPAKTTAV 904
>FTK1_RALSO (Q8XRH0) DNA translocase ftsK 1
Length = 959
Score = 55.5 bits (132), Expect = 5e-07
Identities = 36/157 (22%), Positives = 66/157 (41%), Gaps = 8/157 (5%)
Query: 334 PILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPT-SPITIQYNSLTSEP 392
P+ + PA P + P++P S+ +P++ P ++ + +E + + L + P
Sbjct: 268 PVEDAAPAISPAAEPDAPASAPPEPAEPSPPTVDLEAVRQEAEALLAELRGLMTPLAAAP 327
Query: 393 IILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQ 452
+ +PEPEP P +E P P PE +P+ +A+ EA+P A+
Sbjct: 328 VASPEPEPEPEPE-------AETEVTPEAEAEPEAEPEAEAEPEAEAEAEAEAEAEPEAE 380
Query: 453 QVQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSE 489
P+ V + E T+ P P +E +
Sbjct: 381 APAPESVAPALQEAEAATAAEAPLPAPEPAPAIEADD 417
Score = 42.0 bits (97), Expect = 0.006
Identities = 53/211 (25%), Positives = 76/211 (35%), Gaps = 44/211 (20%)
Query: 339 QPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQY------------N 386
QPA QP PP + R S QP + +W +L E P+ Q
Sbjct: 171 QPAWQP--PPRT-RESPPQPGE----IWPLLNAQGRPEMPLPVAAQPAPVPVPAPAATPK 223
Query: 387 SLTSEP--------IILDQPEPEPRPSDH----SVPRASERPALPSPRRYPRPRPERLVD 434
+ T P I+ P P+PSD S P A + P+ P P P +
Sbjct: 224 AATQAPSSRSALRATIVSSPFHRPQPSDGDQPPSSPEADDAPSAPVEDAAPAISP--AAE 281
Query: 435 PDHPIQADPLHEADPLAQQVQPDQVQQEP----------IQPEPETSVSNHSSVRSPHPQ 484
PD P A P A+P V + V+QE + P V++ P P+
Sbjct: 282 PDAPASAPP-EPAEPSPPTVDLEAVRQEAEALLAELRGLMTPLAAAPVASPEPEPEPEPE 340
Query: 485 VETSEPNLGTSEPNVQTFDVGSPQGASEANS 515
ET +EP + P+ +EA +
Sbjct: 341 AETEVTPEAEAEPEAEPEAEAEPEAEAEAEA 371
Score = 35.8 bits (81), Expect = 0.42
Identities = 40/187 (21%), Positives = 74/187 (39%), Gaps = 24/187 (12%)
Query: 254 PSDGDAPPPKKKKKQVR---IVEKPTRVEPAAQVIRMKTSA---------RVTRSAVQSS 301
PSDGD PP + + + + PAA+ ++ V AV+
Sbjct: 249 PSDGDQPPSSPEADDAPSAPVEDAAPAISPAAEPDAPASAPPEPAEPSPPTVDLEAVRQE 308
Query: 302 SKCVVTESNDDLNSLDALPISTLLNR------TATQLTPILEFQPAEQPTSPPNSPRSSF 355
++ ++ E + L A P+++ T++TP E +P +P + +
Sbjct: 309 AEALLAELRGLMTPLAAAPVASPEPEPEPEPEAETEVTPEAEAEPEAEPEAEAEPEAEA- 367
Query: 356 FQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPEPRPSDHSVPRASER 415
++ EA + P P E +P + + T+ L P PEP P+ + A
Sbjct: 368 --EAEAEAEAEPEAEAPAP-ESVAPALQEAEAATAAEAPL--PAPEPAPAIEADDAAPPP 422
Query: 416 PALPSPR 422
PA+P+ +
Sbjct: 423 PAVPAQK 429
>TONB_SERMA (P26185) TonB protein
Length = 247
Score = 52.8 bits (125), Expect = 3e-06
Identities = 36/129 (27%), Positives = 56/129 (42%), Gaps = 9/129 (6%)
Query: 358 PSQQEAPLWNML-QTPRPSEPTSPITIQYNSLTSEPIILDQPEPEPRPSDHSVPRA--SE 414
P ++AP+ M+ T +EP P + EP +PEPEP P P+A
Sbjct: 42 PKPEDAPISVMMVNTAAMAEPPPPAPAEPEPPQVEP----EPEPEPEPIVEPPPKAIVKP 97
Query: 415 RPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEPIQPEPETSVSN 474
P P P+ P+P+ E+ V P+ P + +P E P + + P++ P V
Sbjct: 98 EPVKPKPKPKPKPKVEKQVKPE-PKKVEP-REPSPFNNDSPAKPIDKAPVKQAPAAPVQG 155
Query: 475 HSSVRSPHP 483
+S P P
Sbjct: 156 NSREVGPRP 164
>NFM_CHICK (P16053) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium
polypeptide) (NF-M)
Length = 857
Score = 52.8 bits (125), Expect = 3e-06
Identities = 58/281 (20%), Positives = 105/281 (36%), Gaps = 27/281 (9%)
Query: 180 LFIESKFIEDLIKAGCTED--------LSTIVSDVFTSTSLKKMGLVKKKVAPAHEDTSD 231
L ++ KF+E++I+ ED LS I ++ ++ K + E+
Sbjct: 442 LKVQHKFVEEIIEETKVEDEKSEMEDALSAIAEEMAAKAQEEEQEEEKAEEEAVEEEAVS 501
Query: 232 EKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSA 291
EK ++A + + K EE + + + DA KK+ I EK E + K A
Sbjct: 502 EKAAEQAAEEEEKEEEEAEEEEAAKSDAAEEGGSKKE-EIEEKEEGEEAEEEEAEAKGKA 560
Query: 292 RVTRSAVQ------------SSSKCVVTESNDDLNSLDA-----LPISTLLNRTATQLTP 334
+ V+ S K VTE + A + A +
Sbjct: 561 EEAGAKVEKVKSPPAKSPPKSPPKSPVTEQAKAVQKAAAEVGKDQKAEKAAEKAAKEEKA 620
Query: 335 ILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTP-RPSEPTSPITIQYNSLTSEPI 393
+PA + P P + P+ ++A +++P +P+ P ++ + + +P
Sbjct: 621 ASPEKPATPKVTSPEKPATPEKPPTPEKAITPEKVRSPEKPTTPEKVVSPEKPASPEKPR 680
Query: 394 ILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVD 434
++P +P+ PR E+PA P R P L D
Sbjct: 681 TPEKPASPEKPATPEKPRTPEKPATPEKPRSPEKPSSPLKD 721
>SSP2_PLAYO (Q01443) Sporozoite surface protein 2 precursor
Length = 827
Score = 52.4 bits (124), Expect = 4e-06
Identities = 69/309 (22%), Positives = 109/309 (34%), Gaps = 23/309 (7%)
Query: 251 PDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESN 310
P+ PS+ + P PKK+ + R KP +P + + S N
Sbjct: 381 PNDPSNPNNPNPKKRNPKRRNPNKPKPNKPNPNKPNPNEPSNPNKPNPNEPSNPNKPNPN 440
Query: 311 DDLNSLDALPISTLLNRTATQLTPILEFQPA--EQPTSP-----PNSPRSSFFQPSQQEA 363
+ N P P+ +P+ +P++P PN P + + E
Sbjct: 441 EPSNPNKPNPNEPSNPNKPNPNEPLNPNEPSNPNEPSNPNAPSNPNEPSNPNEPSNPNEP 500
Query: 364 PLWNMLQTP-RPSEPTSPITIQYNSLTSEPIILDQPEPEPRPSDHSVPRASERPALP--- 419
N P PS P P S +EP+ ++P PS+ + P E P+ P
Sbjct: 501 SNPNEPSNPNEPSNPKKPSNPNEPSNPNEPLNPNEPSNPNEPSNPNEPSNPEEPSNPKEP 560
Query: 420 ----SPRRYPRPRPERLVDPDHPIQ-ADPLH--EADPLAQQVQPDQVQQEPIQPEPETSV 472
P P PE +P P +P++ E +P + +EPI PE
Sbjct: 561 SNPNEPSNPEEPNPEEPSNPKEPSNPEEPINPEELNPKEPSNPEESNPKEPINPEESNPK 620
Query: 473 SNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPTSPETNLSIVPYT 532
+ + +P + EP ++ NV PQ +N P++ N S
Sbjct: 621 EPINPEDNENPLIIQDEPIEPRNDSNVIPILPIIPQ-----KGNNIPSNLPENPSDSEVE 675
Query: 533 YLRPNSLSE 541
Y RPN E
Sbjct: 676 YPRPNDNGE 684
Score = 46.6 bits (109), Expect = 2e-04
Identities = 44/183 (24%), Positives = 75/183 (40%), Gaps = 26/183 (14%)
Query: 344 PTSP--PNSPRSSFFQPSQQEAPLWNMLQTPR-PSEPTSPITIQYNSLTSEPIILDQPEP 400
P +P PN+P + P+ P N P PS P +P + N P ++P+P
Sbjct: 357 PNNPNNPNNPNN----PNNPNNP--NNPNNPNDPSNPNNPNPKKRNPKRRNP---NKPKP 407
Query: 401 EPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQ 460
+ P P P+P P +P +P + +P ++P + P+
Sbjct: 408 NKPNPNKPNPNEPSNPNKPNPNEPSNPNKPNPNEPSNPNKPNPNEPSNP--NKPNPN--- 462
Query: 461 QEPIQPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPT 520
EP+ P ++ + S+ +P E S PN S PN P +E ++ N P+
Sbjct: 463 -EPLNPNEPSNPNEPSNPNAPSNPNEPSNPN-EPSNPN-------EPSNPNEPSNPNEPS 513
Query: 521 SPE 523
+P+
Sbjct: 514 NPK 516
Score = 34.3 bits (77), Expect = 1.2
Identities = 22/107 (20%), Positives = 46/107 (42%), Gaps = 6/107 (5%)
Query: 416 PALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEPIQPEPETSVSNH 475
P +P P P PE+ +P+ P+ + ++ + P+ P P + +N
Sbjct: 281 PQIPIPPVIPNKIPEKPSNPEEPVNPNDPNDPNNPNNPNNPNN-PNNPNNPNNPNNPNNP 339
Query: 476 SSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPTSP 522
++ +P+ + PN + PN + +P + N+ N+P +P
Sbjct: 340 NNPNNPNNPNNPNNPN-NPNNPN----NPNNPNNPNNPNNPNNPNNP 381
>EP40_HUMAN (Q96L91) E1A binding protein p400 (EC 3.6.1.-) (p400 kDa
SWI2/SNF2-related protein) (Domino homolog) (hDomino)
(CAG repeat protein 32) (Trinucleotide repeat-containing
gene 12 protein)
Length = 3160
Score = 52.4 bits (124), Expect = 4e-06
Identities = 87/374 (23%), Positives = 142/374 (37%), Gaps = 48/374 (12%)
Query: 334 PILEFQPAEQPTSPPNSPRSSFF--QPSQQE------APLWNM--LQTPRPSEP--TSPI 381
P+ + P Q PP + + SQQ+ P+ N L TP P P P
Sbjct: 541 PLQQLMPTAQGGMPPTPQAAQLAGQRQSQQQYDPSTGPPVQNAASLHTPLPQLPGRLPPA 600
Query: 382 TIQYNSLTSEPIILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQA 441
+ +L+S QP+ + +P +++P +P P P + + P P Q
Sbjct: 601 GVPTAALSSALQFAQQPQVVEAQTQLQIPVKTQQPNVPIPAP---PSSQLPIPPSQPAQL 657
Query: 442 DPLHEADPLAQQVQPDQVQQEPI----------------QPEPETSVSNHSSVRS--PHP 483
LH P QVQ Q+ P +P P +S S+ + V P P
Sbjct: 658 -ALHVPTPGKVQVQASQLSSLPQMVASTRLPVDPAPPCPRPLPTSSTSSLAPVSGSGPGP 716
Query: 484 QVETSEP-NLGTSEPNVQTFDVGSPQGASEANSSNHPTSPETNLSIVPYTYLRPNSLSEC 542
S P N +S N V S A++ P SP N + + ++L+E
Sbjct: 717 SPARSSPVNRPSSATNKALSPVTSRTPGVVASAPTKPQSPAQNATSSQDS--SQDTLTEQ 774
Query: 543 ISVFN--HEASLMIRNV----QGHTDLSENSDSVAEEWNRLSTWLVAQVPVLMSHLTAER 596
I++ N H+ +R Q + + W+ +L+ ++ + + ER
Sbjct: 775 ITLENQVHQRIAELRKAGLWSQRRLPKLQEAPRPKSHWD----YLLEEMQWMATDFAQER 830
Query: 597 DQRIEAARQRFNRRVAMHEQQQRTKLLEAVEEARRKKEQAEEAARQEAAQAENARLEAE- 655
++ AA++ V HE++Q + EE R + A AR+ N E
Sbjct: 831 RWKVAAAKKLVRTVVRHHEEKQLREERGKKEEQSRLRRIAASTAREIECFWSNIEQVVEI 890
Query: 656 RLEVEAEALRGQAL 669
+L VE E R +AL
Sbjct: 891 KLRVELEEKRKKAL 904
Score = 32.0 bits (71), Expect = 6.0
Identities = 22/89 (24%), Positives = 35/89 (38%), Gaps = 15/89 (16%)
Query: 391 EPIILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPL 450
+P + P P+P+P P P++ P P P+ P P + P
Sbjct: 2537 QPAVAQPPPPQPQPP-------------PPPQQPPPPLPQPQAAGSQPPAGPPAVQPQPQ 2583
Query: 451 AQ-QVQPDQVQQEPIQPEPETSVSNHSSV 478
Q Q QP V Q P + +P + ++V
Sbjct: 2584 PQPQTQPQPV-QAPAKAQPAITTGGSAAV 2611
>MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia
protein 2 (ALL1-related protein)
Length = 5262
Score = 52.0 bits (123), Expect = 6e-06
Identities = 51/190 (26%), Positives = 70/190 (36%), Gaps = 23/190 (12%)
Query: 316 LDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQ-------EAPLWNM 368
L A P+ L+ + L P E P SPP P SS F P ++ E+P
Sbjct: 474 LPASPLPEALHLSRPLEESPLSPPPEESPLSPP--PESSPFSPLEESPLSPPEESPPSPA 531
Query: 369 LQTP-RPSEPTSPITIQYNSLTSEPIILDQPEPEPRPSDHSVPRASERPALPSPRRYPR- 426
L+TP P SP++ + P + P P + P E P P P P
Sbjct: 532 LETPLSPPPEASPLSPPFEESPLSPPPEELPTSPPPEASRLSPPPEESPMSPPPEESPMS 591
Query: 427 --PRPERLVDPDHPIQADPLHEADPLAQQVQPDQV----QQEPIQPEPETSVSNHSSVRS 480
P RL P P E PL+ + ++ + P+ P PE S S
Sbjct: 592 PPPEASRLFPPFEESPLSPPPEESPLSPPPEASRLSPPPEDSPMSPPPE------ESPMS 645
Query: 481 PHPQVETSEP 490
P P+V P
Sbjct: 646 PPPEVSRLSP 655
Score = 45.8 bits (107), Expect = 4e-04
Identities = 47/185 (25%), Positives = 74/185 (39%), Gaps = 16/185 (8%)
Query: 325 LNRTATQLTPILE-FQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITI 383
+ A Q TP+ F P E +P + P + + + Q P + + +P EP P+
Sbjct: 358 IRSVAEQHTPVCSRFSPPEPGDTPTDEPDALYV--ACQGQPKGGHVTSMQPKEP-GPLQC 414
Query: 384 QYNSLTSEPIILDQPEPEPRPSDHSV-PRASERPALPSPRRYPR---PRPERLVDPDHPI 439
+ L + L+ P + + P E P P P P P RL P +
Sbjct: 415 EAKPLGKAGVQLEPQLEAPLNEEMPLLPPPEESPLSPPPEESPTSPPPEASRLSPPPEEL 474
Query: 440 QADPLHEADPLAQQVQPDQVQQEPIQPEPETSVSN---HSSVRSPHPQVETSEPNLGTSE 496
A PL EA L++ +++ P+ P PE S + SS SP + S P
Sbjct: 475 PASPLPEALHLSR-----PLEESPLSPPPEESPLSPPPESSPFSPLEESPLSPPEESPPS 529
Query: 497 PNVQT 501
P ++T
Sbjct: 530 PALET 534
Score = 34.7 bits (78), Expect = 0.93
Identities = 39/132 (29%), Positives = 50/132 (37%), Gaps = 24/132 (18%)
Query: 333 TPILEFQPAEQPT------SPPNSPRSSFFQPSQQEAPLWNMLQTPRPS--EPTSPITIQ 384
+P L +P E P SPP+ P F+P P TPRP P S +
Sbjct: 2067 SPGLGLRPQEPPPAQALAPSPPSHP--DIFRPGSYTDPYAQPPLTPRPQPPPPESCCALP 2124
Query: 385 YNSLTSEPIILDQPEPEPRPSDHS--VPR---------ASERPALPSPRRYPRP--RPER 431
SL S+P P+ + S S PR + P SP Y RP RP+
Sbjct: 2125 PRSLPSDPFSRVPVSPQSQSSSQSPLTPRPLSAEAFCPSPVTPRFQSPDPYSRPPSRPQS 2184
Query: 432 LVDPDHPIQADP 443
DP P+ P
Sbjct: 2185 R-DPFAPLHKPP 2195
Score = 34.7 bits (78), Expect = 0.93
Identities = 36/130 (27%), Positives = 47/130 (35%), Gaps = 13/130 (10%)
Query: 311 DDLNSLDALPISTLLNRTATQLTPILEFQPA-EQPTSPPNSPRSSFFQPSQQEAPLWNML 369
D L DA+ L R L+ + + PA E PT + S Q AP
Sbjct: 4580 DWLKQFDAVLAGYTLKRQLDILSLLKQESPAPEPPTQHRYTYNVSNLDVRQLSAP----- 4634
Query: 370 QTPRPSEPTSPITIQYNSLTSEPII-------LDQPEPEPRPSDHSVPRASERPALPSPR 422
PS P SP+ S +EP++ + P P P P S A +P P
Sbjct: 4635 PPEEPSPPPSPLAPSPASPPTEPLVELPTEPLAEPPVPSPLPLASSPESARPKPRARPPE 4694
Query: 423 RYPRPRPERL 432
RP RL
Sbjct: 4695 EGEDTRPPRL 4704
Score = 34.3 bits (77), Expect = 1.2
Identities = 46/199 (23%), Positives = 70/199 (35%), Gaps = 18/199 (9%)
Query: 348 PNSPRSSFFQPSQ-------QEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEP 400
P +PR+S +P QE P L PS P Y ++P + +P+P
Sbjct: 2055 PLTPRASQVEPQSPGLGLRPQEPPPAQALAPSPPSHPDIFRPGSYTDPYAQPPLTPRPQP 2114
Query: 401 EPRPSDHSVPRASERPALPS-PRRYPRPRPERLVDPDHPIQADPLHE----ADPLAQQVQ 455
P S ++P S LPS P P+ P+ PL P+ + Q
Sbjct: 2115 PPPESCCALPPRS----LPSDPFSRVPVSPQSQSSSQSPLTPRPLSAEAFCPSPVTPRFQ 2170
Query: 456 -PDQVQQEPIQPEPETSVSN-HSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEA 513
PD + P +P+ + H R P+V +L + F P G +
Sbjct: 2171 SPDPYSRPPSRPQSRDPFAPLHKPPRPQPPEVAFKAGSLAHTSLGAGGFPAALPAGPAGE 2230
Query: 514 NSSNHPTSPETNLSIVPYT 532
+ P+ N P T
Sbjct: 2231 LHAKVPSGQPPNFVRSPGT 2249
Score = 34.3 bits (77), Expect = 1.2
Identities = 44/195 (22%), Positives = 63/195 (31%), Gaps = 32/195 (16%)
Query: 316 LDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPS 375
L + I + + QL +L P + SP + +Q + QTP
Sbjct: 3910 LPGVGIMPTVGQLRAQLQGVLAKNPQLRHLSPQQQQQLQALLMQRQLQQSQAVRQTPPYQ 3969
Query: 376 EPTSPITIQYNSLTSEPIILDQPEPEPRPSDH--SVPRASERPALPSPRR---------- 423
EP + + L +P + P P+ P + PR P LP+P
Sbjct: 3970 EPGTQTSPLQGLLGCQPQLGGFPGPQTGPLQELGAGPRPQGPPRLPAPPGALSTGPVLGP 4029
Query: 424 -YPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEPIQPEPETSVSNHSSVRSPH 482
+P P P +P P Q P P + Q P P +P
Sbjct: 4030 VHPTPPPSSPQEPKRPSQL-------PSPSSQLPTEAQLPPTHP------------GTPK 4070
Query: 483 PQVETSEPNLGTSEP 497
PQ T EP G P
Sbjct: 4071 PQGPTLEPPPGRVSP 4085
>MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2)
Length = 5179
Score = 50.8 bits (120), Expect = 1e-05
Identities = 60/298 (20%), Positives = 94/298 (31%), Gaps = 14/298 (4%)
Query: 249 TKPDAPSDGDAPPPKK----------KKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSA- 297
T P P+ +PPP P P+ + T T +
Sbjct: 1397 TTPSPPTTTPSPPPTTTTTLPPTTTPSPPTTTTTTPPPTTTPSPPITTTTTPLPTTTPSP 1456
Query: 298 -VQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSF- 355
+ +++ T + + + P +T T T TP P+ T+P P S+
Sbjct: 1457 PISTTTTPPPTTTPSPPTTTPSPPTTTPSPPTTTTTTPPPTTTPSPPMTTPITPPASTTT 1516
Query: 356 FQPSQQEAPLWNMLQTPRPSE-PTSPITIQYNSLTSEPIILDQPEPEPRPSDHSVPRASE 414
P+ +P TP P+ P+ P T TS + P P P+ + P +
Sbjct: 1517 LPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTT 1576
Query: 415 RPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEPIQPEPETSVSN 474
P+ P+ P P + P P PI P T+
Sbjct: 1577 TPSPPTTTTPSPPTITTTTPPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLP 1636
Query: 475 HSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPTSPETNLSIVPYT 532
++ SP P T+ P T P T + + P+SP T P T
Sbjct: 1637 PTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITTTPSPPTT 1694
Score = 46.6 bits (109), Expect = 2e-04
Identities = 63/306 (20%), Positives = 98/306 (31%), Gaps = 27/306 (8%)
Query: 248 STKPDAP-SDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVV 306
+T P P S PPP P P+ T+ T + S +
Sbjct: 1451 TTTPSPPISTTTTPPPTTTPSPPTTTPSPPTTTPSPP-----TTTTTTPPPTTTPSPPMT 1505
Query: 307 TESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSF-FQPSQQEAPL 365
T ++ P +T T T TP P+ T+P P S+ P+ +P
Sbjct: 1506 TPITPPASTTTLPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPP 1565
Query: 366 WNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPEPRP--SDHSVPRASERPALPS--- 420
TP P+ SP T S + P P P + + P + P+ P+
Sbjct: 1566 PTTTTTPPPTTTPSPPTTTTPSPPTITTTTPPPTTTPSPPTTTTTTPPPTTTPSPPTTTP 1625
Query: 421 -----------PRRYPRPRPERLVDPDHPIQADP---LHEADPLAQQVQPDQVQQEPIQP 466
P P P P P P + P+ P P P
Sbjct: 1626 ITPPTSTTTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPITTTTTPPPTTT-PSSP 1684
Query: 467 EPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPTSPETNL 526
T +++ +P P S P T+ P+ T P + + + P+ P T +
Sbjct: 1685 ITTTPSPPTTTMTTPSPTTTPSSPITTTTTPSSTTTPSPPPTTMTTPSPTTTPSPPTTTM 1744
Query: 527 SIVPYT 532
+ +P T
Sbjct: 1745 TTLPPT 1750
Score = 35.0 bits (79), Expect = 0.71
Identities = 39/190 (20%), Positives = 61/190 (31%), Gaps = 6/190 (3%)
Query: 248 STKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVT 307
+T P P+ PP T P T+ +T ++ T
Sbjct: 1583 TTTPSPPTITTTTPPPTTTPSPPTTT--TTTPPPTTTPSPPTTTPITPPTSTTTLPPTTT 1640
Query: 308 ESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWN 367
S + P +T T T +P + PT+ P+SP ++ PS +
Sbjct: 1641 PSPPPTTTTTPPPTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITT--TPSPPTTTMTT 1698
Query: 368 MLQTPRPSEPTSPITIQYNSLTSE--PIILDQPEPEPRPSDHSVPRASERPALPSPRRYP 425
T PS P + T ++ T P + P P PS + + P S
Sbjct: 1699 PSPTTTPSSPITTTTTPSSTTTPSPPPTTMTTPSPTTTPSPPTTTMTTLPPTTTSSPLTT 1758
Query: 426 RPRPERLVDP 435
P P + P
Sbjct: 1759 TPLPPSITPP 1768
Score = 33.1 bits (74), Expect = 2.7
Identities = 38/178 (21%), Positives = 57/178 (31%), Gaps = 17/178 (9%)
Query: 248 STKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVT 307
+T P P+ PP T + P T+ S T
Sbjct: 1615 TTTPSPPTTTPITPPTST----------TTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTT 1664
Query: 308 ESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAP--- 364
S + P +T + T +P PT+ P+SP ++ PS P
Sbjct: 1665 PSPPITTTTTPPPTTTPSSPITTTPSPPTTTMTTPSPTTTPSSPITTTTTPSSTTTPSPP 1724
Query: 365 ---LWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPEPRPSDHSVPRASERPALP 419
+ T PS PT+ +T + TS P+ P P S P ++ P P
Sbjct: 1725 PTTMTTPSPTTTPSPPTTTMTTLPPTTTSSPLTTTPLPPSITPPTFS-PFSTTTPTTP 1781
>BRD4_HUMAN (O60885) Bromodomain-containing protein 4 (HUNK1 protein)
Length = 1362
Score = 50.8 bits (120), Expect = 1e-05
Identities = 90/408 (22%), Positives = 141/408 (34%), Gaps = 97/408 (23%)
Query: 330 TQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLT 389
T L P ++ Q P PP P S Q QQ+ P P P +P P Q+
Sbjct: 942 TPLLPSVKVQSQPPPPLPP-PPHPSVQQQLQQQPP------PPPPPQPQPPPQQQH---- 990
Query: 390 SEPIILDQPEPEP---RPSDHSV------PRASERPALPSPRRYPRP----RPERLVDPD 436
QP P P +P S P ++P P P + P P +P++++
Sbjct: 991 -------QPPPRPVHLQPMQFSTHIQQPPPPQGQQPPHPPPGQQPPPPQPAKPQQVIQHH 1043
Query: 437 H--------PIQADPLHEA--------------------DPLAQQVQPDQVQQEP---IQ 465
H P L EA P Q VQP + + +Q
Sbjct: 1044 HSPRHHKSDPYSTGHLREAPSPLMIHSPQMSQFQSLTHQSPPQQNVQPKKQELRAASVVQ 1103
Query: 466 PEPETSVSNHSSVRSPHPQVETSEPNLGTS----------------EPNVQTFDVGSPQG 509
P+P V + SP + E P+L P ++ DVG P
Sbjct: 1104 PQPLVVVKEEK-IHSPIIRSEPFSPSLRPEPPKHPESIKAPVHLPQRPEMKPVDVGRPVI 1162
Query: 510 ASEANSSNHPTSP-------ETNLSIVPYTYLRPNSLSECISVFNHEASLMIRNVQGHTD 562
++ P +P E + P L+ ++ S+ + + +D
Sbjct: 1163 RPPEQNAPPPGAPDKDKQKQEPKTPVAPKKDLKIKNMGSWASLVQKHPTTPSSTAKSSSD 1222
Query: 563 LSENSDSVAEEWNRLSTWLVAQVPVLMSHLTAERDQRIEAARQRFNRRVAMHEQQQRTKL 622
E A E L AQ H E++ R+ R R EQ +R
Sbjct: 1223 SFEQFRRAAREKEEREKALKAQA----EHAEKEKE-RLRQERMRSREDEDALEQARR--- 1274
Query: 623 LEAVEEARRKKEQAEEAAR-QEAAQAENARLEAERLEVEAEALRGQAL 669
A EEARR++EQ ++ + Q+ Q + A A +A++ + Q++
Sbjct: 1275 --AHEEARRRQEQQQQQRQEQQQQQQQQAAAVAAAATPQAQSSQPQSM 1320
Score = 46.6 bits (109), Expect = 2e-04
Identities = 55/209 (26%), Positives = 80/209 (37%), Gaps = 31/209 (14%)
Query: 333 TPILEFQPAEQPTS-------PPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQY 385
TP+L P QP PP P +S Q LQ +P P P
Sbjct: 897 TPLLPQPPMAQPPQVLLEDEEPPAPPLTSM-----QMQLYLQQLQKVQPPTPLLPSV--- 948
Query: 386 NSLTSEPIILDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERL-VDPDHPIQADPL 444
+ S+P P P P P SV + ++ P P P+P P++ P P+ P+
Sbjct: 949 -KVQSQP-----PPPLPPPPHPSVQQQLQQQPPPPPPPQPQPPPQQQHQPPPRPVHLQPM 1002
Query: 445 HEADPLAQQVQPDQVQQEP-----IQPEPETSVSNHSSVRSPH-PQVETSEP-NLGTSEP 497
+ + QQ P Q QQ P QP P ++ H P+ S+P + G
Sbjct: 1003 QFSTHI-QQPPPPQGQQPPHPPPGQQPPPPQPAKPQQVIQHHHSPRHHKSDPYSTGHLRE 1061
Query: 498 NVQTFDVGSPQGASEANSSNHPTSPETNL 526
+ SPQ S+ S H + P+ N+
Sbjct: 1062 APSPLMIHSPQ-MSQFQSLTHQSPPQQNV 1089
Score = 41.6 bits (96), Expect = 0.008
Identities = 68/283 (24%), Positives = 106/283 (37%), Gaps = 26/283 (9%)
Query: 232 EKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSA 291
EK KK+ K K K +E + + S PPPKK KK ++ EPA MK+
Sbjct: 540 EKDKKEKKKEKHKRKEEVEENKKSKAKEPPPKKTKKNNSSNSNVSKKEPAP----MKSKP 595
Query: 292 RVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSP 351
T + + KC S ++ L +L I+ L ++ I++ + S P+
Sbjct: 596 PPTYES-EEEDKC-KPMSYEEKRQL-SLDINKLPGEKLGRVVHIIQSREPSLKNSNPDEI 652
Query: 352 RSSF--FQPS---QQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPEPRPSD 406
F +PS + E + + L+ R + + + S + + E S
Sbjct: 653 EIDFETLKPSTLRELERYVTSCLRKKRKPQ-AEKVDVIAGSSKMKGFSSSESESSSE-SS 710
Query: 407 HSVPRASERPALPSPRRYPRPRPERLVDPDHPIQ------ADPLHEADPLAQQ---VQPD 457
S SE P ++ P E+ H Q A + P QQ P
Sbjct: 711 SSDSEDSETEMAPKSKKKGHPGREQKKHHHHHHQQMQQAPAPVPQQPPPPPQQPPPPPPP 770
Query: 458 QVQQEPIQPEPETSVSNHSS---VRSPHPQVETSEPNLGTSEP 497
Q QQ+P P P S+ ++ SP P + T P L P
Sbjct: 771 QQQQQPPPPPPPPSMPQQAAPAMKSSPPPFIATQVPVLEPQLP 813
Score = 39.7 bits (91), Expect = 0.029
Identities = 44/162 (27%), Positives = 62/162 (38%), Gaps = 24/162 (14%)
Query: 340 PAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPI------ 393
P +Q PP P S QQ AP P + + Q +PI
Sbjct: 770 PQQQQQPPPPPPPPSM---PQQAAPAMKSSPPPFIATQVPVLEPQLPGSVFDPIGHFTQP 826
Query: 394 ILDQPEPE-----PRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEAD 448
IL P+PE P+P +HS P + A+ SP P++ P + A P A
Sbjct: 827 ILHLPQPELPPHLPQPPEHSTPPHLNQHAVVSPPALHNALPQQPSRPSNRAAALPPKPAR 886
Query: 449 PLAQQVQPDQVQQEPIQPEPETSVSNHSSVRSPHPQVETSEP 490
P A V P + Q P+ P+P + + P +E EP
Sbjct: 887 PPA--VSP-ALTQTPLLPQPPMA-------QPPQVLLEDEEP 918
Score = 37.7 bits (86), Expect = 0.11
Identities = 42/177 (23%), Positives = 67/177 (37%), Gaps = 25/177 (14%)
Query: 321 ISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSP 380
+ST+ N T P + QP PP F + + + T P +P
Sbjct: 193 VSTVPNTTQASTPPQTQ---TPQPNPPPVQATPHPFPAVTPDLIVQTPVMTVVPPQP--- 246
Query: 381 ITIQYNSLTSEPIILDQPEPEPRPSDHSVPRASERP---ALPSPRRYPRPRPERLVDPDH 437
L + P + QP+P P P+ P S P A P P + + +R D
Sbjct: 247 -------LQTPPPVPPQPQPPPAPAPQ--PVQSHPPIIAATPQPVK-TKKGVKRKADTTT 296
Query: 438 PIQADPLHEADPLAQQVQPDQVQQ-----EPIQPEPETSVSNHSSVRSPHPQVETSE 489
P DP+HE L + + ++ Q P++P P+ V + +P + SE
Sbjct: 297 PTTIDPIHEPPSLPPEPKTTKLGQRRESSRPVKP-PKKDVPDSQQHPAPEKSSKVSE 352
>UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89
(Uncoordinated protein 89)
Length = 6632
Score = 50.1 bits (118), Expect = 2e-05
Identities = 58/298 (19%), Positives = 109/298 (36%), Gaps = 14/298 (4%)
Query: 213 KKMGLVKKKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQ-VRI 271
KK +K V + ++ +K DK K ++ P+ + D P KK+K ++
Sbjct: 1603 KKEKSPEKSVVEELKSPKEKSPEKADDKPKSPTKKEKSPEKSATEDVKSPTKKEKSPEKV 1662
Query: 272 VEKPTRVEPAAQVIRMKTSARVTRSAVQSSSKCVVTESNDDLNSLDALPISTLLN--RTA 329
EKPT KT V + S V E + P +++ ++
Sbjct: 1663 EEKPTSPTKKESSPTKKTDDEVKSPTKKEKSPQTVEEKPASPTKKEKSPEKSVVEEVKSP 1722
Query: 330 TQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEAPLWNMLQTPRPSEPTSPITIQYNSLT 389
+ +P + + PT SP S + + + +P SP
Sbjct: 1723 KEKSPEKAEEKPKSPTKKEKSPEKSAAEEVKSPTKKEKSPEKSAEEKPKSP-----TKKE 1777
Query: 390 SEPI-ILDQPEPEPRPSDHSVPRASERPALPSPRRY--PRPRPERLVDP--DHPIQADPL 444
S P+ + D P + S + E+PA P+ + + E L P + P
Sbjct: 1778 SSPVKMADDEVKSPTKKEKSPEKVEEKPASPTKKEKTPEKSAAEELKSPTKKEKSPSSPT 1837
Query: 445 HEADPLAQQVQPDQVQQEPIQPEPETS-VSNHSSVRSPHPQVETSEPNLGTSEPNVQT 501
+ +++ P++ +++P P P+ S + +S P+ E T + +QT
Sbjct: 1838 KKTGDESKEKSPEKPEEKPKSPTPKKSPPGSPKKKKSKSPEAEKPPAPKLTRDLKLQT 1895
Score = 47.8 bits (112), Expect = 1e-04
Identities = 76/376 (20%), Positives = 128/376 (33%), Gaps = 84/376 (22%)
Query: 219 KKKVAPAHEDTSDEKKKKKADKSKRKHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRV 278
KK +P ++ S EK ++K +K E K +P K+K V+ PT+
Sbjct: 1374 KKSPSPDKKEKSPEKTEEKPASPTKKTGEEVKSPKEKSPASPTKKEKSPAAEEVKSPTKK 1433
Query: 279 E--PAAQVIRMKTSARVT---------RSAVQSSSK-------------CVVTESNDDLN 314
E P++ + K+ + T +S +S +K V E + D
Sbjct: 1434 EKSPSSPTKKEKSPSSPTKKTGDEVKEKSPPKSPTKKEKSPEKPEDVKSPVKKEKSPDAT 1493
Query: 315 SLDALPISTLLNRTATQLTPILEFQPAEQPTS------------PPNSPRSSFFQPSQ-- 360
++ + T + +T T +T + + E TS P SP P +
Sbjct: 1494 NIVEVSSETTIEKTETTMTTEMTHESEESRTSVKKEKTPEKVDEKPKSPTKKDKSPEKSI 1553
Query: 361 -----------------QEAPLWNMLQTPRPSEPTSPITIQYNSLTSEPIILDQPEPE-- 401
+E P + P +P SP N + S PE
Sbjct: 1554 TEEIKSPVKKEKSPEKVEEKPASPTKKEKSPEKPASPTKKSENEVKSPTKKEKSPEKSVV 1613
Query: 402 ---PRPSDHSVPRASERPALPSPRRYPRPRPERLVDPDHPIQADPLHEADPLAQQVQPDQ 458
P + S +A ++P P+ + PE+ D P ++ P++
Sbjct: 1614 EELKSPKEKSPEKADDKPKSPTKK---EKSPEKSATED---------VKSPTKKEKSPEK 1661
Query: 459 VQQEPIQP---EPETSVSNHSSVRSP-----HPQVETSEPNLGT----SEPNVQTFDVGS 506
V+++P P E + V+SP PQ +P T S +V S
Sbjct: 1662 VEEKPTSPTKKESSPTKKTDDEVKSPTKKEKSPQTVEEKPASPTKKEKSPEKSVVEEVKS 1721
Query: 507 PQGASEANSSNHPTSP 522
P+ S + P SP
Sbjct: 1722 PKEKSPEKAEEKPKSP 1737
Score = 34.3 bits (77), Expect = 1.2
Identities = 36/201 (17%), Positives = 77/201 (37%), Gaps = 12/201 (5%)
Query: 184 SKFIEDLIKAGCTEDLSTIVSDVFTSTSLKKMGLVKKKVAPAHEDTSDEKKKKKADKSKR 243
+K +D +K+ ++ S + ++ KK +K V + ++ +K +K K
Sbjct: 1677 TKKTDDEVKSPTKKEKSPQTVEEKPASPTKKEKSPEKSVVEEVKSPKEKSPEKAEEKPKS 1736
Query: 244 KHEESTKPDAPSDGDAPPPKKKKKQVRIVEKPTRVEPAAQVIRMKTSARVTRSAVQSSSK 303
++ P+ + + P KK+K EK +P + + + ++ V+S
Sbjct: 1737 PTKKEKSPEKSAAEEVKSPTKKEKS---PEKSAEEKPKSPTKKESSPVKMADDEVKSP-- 1791
Query: 304 CVVTESNDDLNSLDALPISTLLNRTATQLTPILEFQPAEQPTSPPNSPRSSFFQPSQQEA 363
T+ ++ P S + + E + + P+SP S++++
Sbjct: 1792 ---TKKEKSPEKVEEKPASPTKKEKTPEKSAAEELKSPTKKEKSPSSPTKKTGDESKEKS 1848
Query: 364 P----LWNMLQTPRPSEPTSP 380
P TP+ S P SP
Sbjct: 1849 PEKPEEKPKSPTPKKSPPGSP 1869
>Y046_UREPA (Q9PR99) Hypothetical protein UU046
Length = 791
Score = 49.7 bits (117), Expect = 3e-05
Identities = 43/164 (26%), Positives = 68/164 (41%), Gaps = 26/164 (15%)
Query: 382 TIQYNSLTSEPIILDQPEPEPRPSDHSVPRAS--------------ERPALPS--PRRYP 425
+I+ N+L P QP+PEP+P+ P + E P P P P
Sbjct: 65 SIKDNNLKEIPKPKPQPKPEPQPTPFPDPIPTPPKKEELKKPEIKPEEPKKPEIKPEPIP 124
Query: 426 RPRPERLVDPDHPIQADPLHEADPLAQQVQPDQVQQEP-IQPEPETSVSNHSSVRSPHPQ 484
+P+P+ + P P++ P E P + ++ + EP QP+P+ + N S+VR
Sbjct: 125 KPKPQPIPQPTPPVETKPKEELLPPSPPPPKEEPKPEPDPQPQPQ-QIPNQSTVRKIELN 183
Query: 485 VETSEPNLGTSEPNVQTFDVGSPQGASEANSSNHPTSPETNLSI 528
+ + P QTF G S N P TN+S+
Sbjct: 184 GVLVDAEVEVPPPR-QTFKYDQDNGLSNLN-------PYTNISV 219
Score = 32.3 bits (72), Expect = 4.6
Identities = 24/112 (21%), Positives = 44/112 (38%), Gaps = 9/112 (8%)
Query: 395 LDQPEPEPRPSDHSVPRASERPALPSPRRYPRPRPERLVDP--DHPIQADPLHEADPLAQ 452
LD + P D+++ + P P P P P+ + P ++ + +P
Sbjct: 57 LDYQKARPSIKDNNLKEIPKPKPQPKPEPQPTPFPDPIPTPPKKEELKKPEIKPEEPKKP 116
Query: 453 QVQPDQV---QQEPI-QPEPETSVSNHSSVRSPHPQVETSEPNLGTSEPNVQ 500
+++P+ + + +PI QP P + P P EP EP+ Q
Sbjct: 117 EIKPEPIPKPKPQPIPQPTPPVETKPKEELLPPSPPPPKEEPK---PEPDPQ 165
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.128 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 84,979,632
Number of Sequences: 164201
Number of extensions: 3894444
Number of successful extensions: 25813
Number of sequences better than 10.0: 1008
Number of HSP's better than 10.0 without gapping: 159
Number of HSP's successfully gapped in prelim test: 893
Number of HSP's that attempted gapping in prelim test: 19913
Number of HSP's gapped (non-prelim): 4044
length of query: 734
length of database: 59,974,054
effective HSP length: 118
effective length of query: 616
effective length of database: 40,598,336
effective search space: 25008574976
effective search space used: 25008574976
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0134.7


