

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.5
(510 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
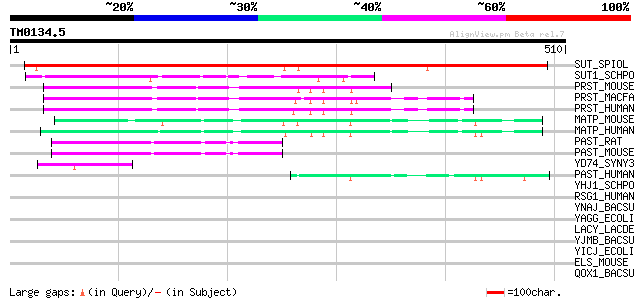
Score E
Sequences producing significant alignments: (bits) Value
SUT_SPIOL (Q03411) Sucrose transport protein (Sucrose permease) ... 639 0.0
SUT1_SCHPO (O14091) General alpha-glucoside permease 137 5e-32
PRST_MOUSE (Q8K0H7) Prostein 117 9e-26
PRST_MACFA (Q95KI5) Prostein (QmoA-10594/QtrA-11310) 115 2e-25
PRST_HUMAN (Q96JT2) Prostein 114 6e-25
MATP_MOUSE (P58355) Membrane-associated transporter protein (AIM... 110 1e-23
MATP_HUMAN (Q9UMX9) Membrane-associated transporter protein (AIM... 103 1e-21
PAST_RAT (Q8K4S3) Proton-associated sugar transporter A (PAST-A) 97 1e-19
PAST_MOUSE (Q8BIV7) Proton-associated sugar transporter A (PAST-... 96 3e-19
YD74_SYNY3 (P74168) Hypothetical symporter sll1374 47 2e-04
PAST_HUMAN (Q9Y2W3) Proton-associated sugar transporter A (PAST-... 45 4e-04
YHJ1_SCHPO (Q9C0V8) Probable transporter PB10D8.01 40 0.014
RSG1_HUMAN (P20936) Ras GTPase-activating protein 1 (GTPase-acti... 39 0.032
YNAJ_BACSU (P94488) Hypothetical symporter ynaJ 39 0.042
YAGG_ECOLI (P75683) Hypothetical symporter yagG 39 0.042
LACY_LACDE (P22733) Lactose permease (Lactose-proton symporter) ... 39 0.042
YJMB_BACSU (O34961) Hypothetical symporter yjmB 37 0.12
YICJ_ECOLI (P31435) Hypothetical symporter yicJ 37 0.12
ELS_MOUSE (P54320) Elastin precursor (Tropoelastin) 36 0.27
QOX1_BACSU (P34956) Quinol oxidase polypeptide I (EC 1.10.3.-) (... 35 0.35
>SUT_SPIOL (Q03411) Sucrose transport protein (Sucrose permease)
(Sucrose-proton symporter)
Length = 525
Score = 639 bits (1648), Expect = 0.0
Identities = 319/499 (63%), Positives = 388/499 (76%), Gaps = 18/499 (3%)
Query: 14 GSSLQIEAHI---PQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASF 70
GSSL +E + P+ + L+K+ VAS+AAG+QFGWALQLSLLTPYVQ LG+PH WA++
Sbjct: 16 GSSLHLEKNPTTPPEAEATLKKLGLVASVAAGVQFGWALQLSLLTPYVQLLGIPHTWAAY 75
Query: 71 IWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGD 130
IWLCGP+SG++VQP+VGYYSDR T RFGRRRPFI AGA VA++V LIG+AADIG + GD
Sbjct: 76 IWLCGPISGMIVQPLVGYYSDRCTSRFGRRRPFIAAGAALVAVAVGLIGFAADIGAASGD 135
Query: 131 DLAKKTRPRAVAIFVFGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAV 190
+PRA+A+FV GFWILDVANN LQGPCRA L D+AAG Q KTR A FFSFFMA+
Sbjct: 136 PTGNVAKPRAIAVFVVGFWILDVANNTLQGPCRALLADMAAGSQTKTRYANAFFSFFMAL 195
Query: 191 GNVLGYAAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPL 250
GN+ GYAAG+YS L+ +FPFT T AC +CANLKSCFF SI LL++L+I AL V++ +
Sbjct: 196 GNIGGYAAGSYSRLYTVFPFTKTAACDVYCANLKSCFFISITLLIVLTILALSVVKERQI 255
Query: 251 T------KKPAADVDSPVSC-----FGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLF 299
T ++ + ++ C FG L GA K+L KPM IL+LVTA+NW+AWFPF LF
Sbjct: 256 TIDEIQEEEDLKNRNNSSGCARLPFFGQLIGALKDLPKPMLILLLVTALNWIAWFPFLLF 315
Query: 300 DTDWMGREVYGGVVGE-KAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWG 358
DTDWMG+EVYGG VGE K YD GV AG+LGLMIN+VVL MSL++E L R++GG K LWG
Sbjct: 316 DTDWMGKEVYGGTVGEGKLYDQGVHAGALGLMINSVVLGVMSLSIEGLARMVGGAKRLWG 375
Query: 359 IVNFILAICMAMTVLITKTAEHDR---LISGGATVGAPSSGVKAGAIAFFGVMGIPLAIN 415
IVN ILA+C+AMTVL+TK+AEH R I G A P +GVK GA+A F V+GIPLAI
Sbjct: 376 IVNIILAVCLAMTVLVTKSAEHFRDSHHIMGSAVPPPPPAGVKGGALAIFAVLGIPLAIT 435
Query: 416 FSVPFALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGA 475
FS+PFALASI+S+++G+GQGLSLGVLNLAIVVPQM VS SGPWDA+FGGGNLPAFVVGA
Sbjct: 436 FSIPFALASIFSASSGSGQGLSLGVLNLAIVVPQMFVSVTSGPWDAMFGGGNLPAFVVGA 495
Query: 476 VMAAVSAIMAIVLLPTPKP 494
V A SA+++ LLP+P P
Sbjct: 496 VAATASAVLSFTLLPSPPP 514
>SUT1_SCHPO (O14091) General alpha-glucoside permease
Length = 553
Score = 137 bits (346), Expect = 5e-32
Identities = 100/334 (29%), Positives = 165/334 (48%), Gaps = 35/334 (10%)
Query: 15 SSLQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLC 74
+S + IP +SS +IA+ G+Q W+++L TPY+ +LG+ W S IW+
Sbjct: 20 ASSPFKESIPSRSSLY--LIALTVSLLGVQLTWSVELGYGTPYLFSLGLRKEWTSIIWIA 77
Query: 75 GPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSM--GDDL 132
GP++G+L+QPI G SDR R GRRRPF+L ++ S+FL+G+A DI + + L
Sbjct: 78 GPLTGILIQPIAGILSDRVNSRIGRRRPFMLCASLLGTFSLFLMGWAPDICLFIFSNEVL 137
Query: 133 AKKTRPRAVAIFVFGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGN 192
K+ + + ++LDVA N++ R+ + D DQ+ A + + VGN
Sbjct: 138 MKRV---TIVLATISIYLLDVAVNVVMASTRSLIVDSVRSDQQ--HEANSWAGRMIGVGN 192
Query: 193 VLGYAAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTK 252
VLGY G Y L++IF F +F C SI L+L ++I + +++
Sbjct: 193 VLGYLLG-YLPLYRIFSFL------NFTQLQVFCVLASISLVLTVTITTIF------VSE 239
Query: 253 KPAADVDSPVSCFGDLFGAFKELKKPMWIL-------MLVTAVNWVAWFPFFLFDTDWMG 305
+ V+ S G++F F +++ + L V + WFPF + T ++G
Sbjct: 240 RRFPPVEHEKSVAGEIFEFFTTMRQSITALPFTLKRICFVQFFAYFGWFPFLFYITTYVG 299
Query: 306 ----REVYGGVVGEKAYDDGVRAGSLGLMINAVV 335
R G E+ +D R GS L++ A++
Sbjct: 300 ILYLRHAPKG--HEEDWDMATRQGSFALLLFAII 331
>PRST_MOUSE (Q8K0H7) Prostein
Length = 553
Score = 117 bits (292), Expect = 9e-26
Identities = 93/347 (26%), Positives = 156/347 (44%), Gaps = 40/347 (11%)
Query: 32 KMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSD 91
+++ V + G++ A ++ + P + +GV + + + GPV GL+ P++G SD
Sbjct: 17 QLLLVNLLTFGLEVCLAAGITYVPPLLLEVGVEEKFMTMVLGIGPVLGLVSVPLLGSASD 76
Query: 92 RTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFGFWIL 151
+ R+GRRRPFI A ++ V +S+FLI A + + D TRP +A+ + G +L
Sbjct: 77 QWRGRYGRRRPFIWALSLGVLLSLFLIPRAGWLAGLLYPD----TRPLELALLILGVGLL 132
Query: 152 DVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFT 211
D + P A L DL D R A ++F +++G LGY A
Sbjct: 133 DFCGQVCFTPLEALLSDLFR-DPDHCRQAFSVYAFMISLGGCLGYLLPAID--------W 183
Query: 212 VTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVSC------- 264
T + + C F + L+ ++ +AA ++V + + P VS
Sbjct: 184 DTSVLAPYLGTQEECLFGLLTLIFLICMAATLFVTEEAVLGPPEPAEGLLVSAVSRRCCP 243
Query: 265 ---------FGDLFGAFKEL--KKPMWILMLVTA--VNWVAWFPFFLFDTDWMGREVYGG 311
G LF ++L + P + L A +W+A F LF TD++G +Y G
Sbjct: 244 CHVGLAFRNLGTLFPRLQQLCCRMPRTLRRLFVAELCSWMALMTFTLFYTDFVGEGLYQG 303
Query: 312 V-------VGEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILG 351
V + YD+G+R GSLGL + + SL ++ L + G
Sbjct: 304 VPRAEPGTEARRHYDEGIRMGSLGLFLQCAISLVFSLVMDRLVQKFG 350
>PRST_MACFA (Q95KI5) Prostein (QmoA-10594/QtrA-11310)
Length = 553
Score = 115 bits (289), Expect = 2e-25
Identities = 113/424 (26%), Positives = 186/424 (43%), Gaps = 66/424 (15%)
Query: 32 KMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSD 91
+++ + + G++ A ++ + P + +GV + + + GPV GL+ P++G SD
Sbjct: 17 QLLLINLLTFGLEVCLAAGITYVPPLLLEVGVEEKFMTMVLGIGPVLGLVSVPLLGSASD 76
Query: 92 RTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFGFWIL 151
R+GRRRPFI A ++ + +S+FLI A + + D RP +A+ + G +L
Sbjct: 77 HWRGRYGRRRPFIWALSLGILLSLFLIPRAGWLAGLLCPD----PRPLELALLILGVGLL 132
Query: 152 DVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFT 211
D + P A L DL D R A ++F +++G LGY A
Sbjct: 133 DFCGQVCFTPLEALLSDLFR-DPDHCRQAYSVYAFMISLGGCLGYLLPAID--------W 183
Query: 212 VTDACPSFCANLKSCFFFSILLLLILSIAA-LIYVEDTPL-TKKPAADVDSP------VS 263
T A + + C F + L+ + +AA L+ E+ L +PA + +P
Sbjct: 184 DTSALAPYLGTQEECLFGLLTLIFLTCVAATLLVAEEAALGPAEPAEGLSAPSLPSHCCP 243
Query: 264 CFGDLFGAFKEL------------KKPMWILMLVTA--VNWVAWFPFFLFDTDWMGREVY 309
C+ L AF+ L + P + L A +W+A F LF TD++G +Y
Sbjct: 244 CWARL--AFRNLGALLPRLHQLCCRMPRTLRRLFVAELCSWMALMTFTLFYTDFVGEGLY 301
Query: 310 GGV----VGEKA---YDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNF 362
GV +G +A YD+GVR GSLGL + + SL ++ L + G
Sbjct: 302 QGVPRAELGTEARRHYDEGVRMGSLGLFLQCAISLVFSLVMDRLVQRFG----------- 350
Query: 363 ILAICMAMTVLITKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFAL 422
A+ +A ++ GAT + S V + A G L I +P+ L
Sbjct: 351 TRAVYLASVAAFP--------VAAGATCLSHSVAVVTASAALTGFTFSALQI---LPYTL 399
Query: 423 ASIY 426
AS+Y
Sbjct: 400 ASLY 403
>PRST_HUMAN (Q96JT2) Prostein
Length = 553
Score = 114 bits (285), Expect = 6e-25
Identities = 112/422 (26%), Positives = 181/422 (42%), Gaps = 62/422 (14%)
Query: 32 KMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSD 91
+++ V + G++ A ++ + P + +GV + + + GPV GL+ P++G SD
Sbjct: 17 QLLLVNLLTFGLEVCLAAGITYVPPLLLEVGVEEKFMTMVLGIGPVLGLVCVPLLGSASD 76
Query: 92 RTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFGFWIL 151
R+GRRRPFI A ++ + +S+FLI A + + D RP +A+ + G +L
Sbjct: 77 HWRGRYGRRRPFIWALSLGILLSLFLIPRAGWLAGLLCPD----PRPLELALLILGVGLL 132
Query: 152 DVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFT 211
D + P A L DL D R A ++F +++G LGY A
Sbjct: 133 DFCGQVCFTPLEALLSDLFR-DPDHCRQAYSVYAFMISLGGCLGYLLPAID--------W 183
Query: 212 VTDACPSFCANLKSCFFFSILLLLILSIAA-LIYVEDTPL-TKKPAADVD----SPVSCF 265
T A + + C F + L+ + +AA L+ E+ L +PA + SP C
Sbjct: 184 DTSALAPYLGTQEECLFGLLTLIFLTCVAATLLVAEEAALGPTEPAEGLSAPSLSPHCCP 243
Query: 266 GDLFGAFKEL------------KKPMWILMLVTA--VNWVAWFPFFLFDTDWMGREVYGG 311
AF+ L + P + L A +W+A F LF TD++G +Y G
Sbjct: 244 CRARLAFRNLGALLPRLHQLCCRMPRTLRRLFVAELCSWMALMTFTLFYTDFVGEGLYQG 303
Query: 312 V-------VGEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFIL 364
V + YD+GVR GSLGL + + SL ++ L + G
Sbjct: 304 VPRAEPGTEARRHYDEGVRMGSLGLFLQCAISLVFSLVMDRLVQRFG-----------TR 352
Query: 365 AICMAMTVLITKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALAS 424
A+ +A ++ GAT + S V + A G L I +P+ LAS
Sbjct: 353 AVYLASVAAFP--------VAAGATCLSHSVAVVTASAALTGFTFSALQI---LPYTLAS 401
Query: 425 IY 426
+Y
Sbjct: 402 LY 403
>MATP_MOUSE (P58355) Membrane-associated transporter protein (AIM-1
protein) (Melanoma antigen AIM1) (Underwhite protein)
Length = 530
Score = 110 bits (274), Expect = 1e-23
Identities = 122/521 (23%), Positives = 201/521 (38%), Gaps = 120/521 (23%)
Query: 42 GIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRR 101
G +F +A++ + +TP + ++G+P S +WL P+ G L+QP+VG SD R+GRRR
Sbjct: 44 GREFCYAVEAAYVTPVLLSVGLPKSLYSMVWLLSPILGFLLQPVVGSASDHCRARWGRRR 103
Query: 102 PFILAGAVAVAISVFLIGYAADI-GHSMGDDLAKKTRPR---AVAIFVFGFWILDVANNM 157
P+IL A+ + L+G A + G ++ L R + A++I + G + D + +
Sbjct: 104 PYILTLAI-----MMLLGMALYLNGDAVVSALVANPRQKLIWAISITMVGVVLFDFSADF 158
Query: 158 LQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTVTDACP 217
+ GP +A+L D+ + K+ + + + F G LGY GA +H D
Sbjct: 159 IDGPIKAYLFDVCSHQDKE--KGLHYHALFTGFGGALGYILGAIDWVH-------LDLGR 209
Query: 218 SFCANLKSCFFFSILLLLILSIAALIYVEDTPL--------------------------- 250
+ FFFS L+L++ I L + + PL
Sbjct: 210 LLGTEFQVMFFFSALVLILCFITHLCSIPEAPLRDAATDPPSQQDPQGSSLSASGMHEYG 269
Query: 251 ----TKKPAADVDSPVS---------------CFGDLFGAFKELKKPMWILMLVTAVNWV 291
K AD + PV L A + L + + W
Sbjct: 270 SIEKVKNGGADTEQPVQEWKNKKPSGQSQRTMSMKSLLRALVNMPSHYRCLCVSHLIGWT 329
Query: 292 AWFPFFLFDTDWMGREVYGG-------VVGEKAYDDGVRAGSLGLMINAVVLAFMSLAVE 344
A+ LF TD+MG+ VY G Y+ GV G GL IN+V + S +
Sbjct: 330 AFLSNMLFFTDFMGQIVYHGDPYGAHNSTEFLIYERGVEVGCWGLCINSVFSSVYSYFQK 389
Query: 345 PLGRILGGVKNLWGIVNFILAICMAMTVLITKTAEHDRLISGGATVGA-PSSGVKAGAIA 403
+ + G+K L+ + + + G +G P+ +
Sbjct: 390 AMVSYI-GLKGLYFMGYLLFGL-------------------GTGFIGLFPNVYSTLVLCS 429
Query: 404 FFGVMGIPLAINFSVPFALASIY---------------SSAAGAGQGLSLGVLNLAIVVP 448
FGVM L ++VPF L + Y G G+G+ L + +
Sbjct: 430 MFGVMSSTL---YTVPFNLIAEYHREEEKEKGQEAPGGPDNQGRGKGVDCAALTCMVQLA 486
Query: 449 QMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLL 489
Q+LV GG + G+V+ V A+ L+
Sbjct: 487 QILVG----------GGLGFLVNMAGSVVVVVITASAVSLI 517
>MATP_HUMAN (Q9UMX9) Membrane-associated transporter protein (AIM-1
protein) (Melanoma antigen AIM1)
Length = 530
Score = 103 bits (257), Expect = 1e-21
Identities = 121/530 (22%), Positives = 206/530 (38%), Gaps = 112/530 (21%)
Query: 29 PLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGY 88
P ++I + G +F +A++ + +TP + ++G+P S +W P+ G L+QP+VG
Sbjct: 31 PTSRLIMHSMAMFGREFCYAVEAAYVTPVLLSVGLPSSLYSIVWFLSPILGFLLQPVVGS 90
Query: 89 YSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFGF 148
SD R+GRRRP+IL V + + + L A + ++ + +K A+++ + G
Sbjct: 91 ASDHCRSRWGRRRPYILTLGVMMLVGMALYLNGATVVAALIANPRRKL-VWAISVTMIGV 149
Query: 149 WILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIF 208
+ D A + + GP +A+L D+ + K+ + + + F G LGY GA H
Sbjct: 150 VLFDFAADFIDGPIKAYLFDVCSHQDKE--KGLHYHALFTGFGGALGYLLGAIDWAH--- 204
Query: 209 PFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTK------KPAADVDSPV 262
+ + FFFS L+L + L + + PLT+ D P+
Sbjct: 205 ----LELGRLLGTEFQVMFFFSALVLTLCFTVHLCSISEAPLTEVAKGIPPQQTPQDPPL 260
Query: 263 SCFGDL-FGAFKELK-----------------------KPMWILMLVTA----------- 287
S G +G+ +++K + M + L+ A
Sbjct: 261 SSDGMYEYGSIEKVKNGYVNPELAMQGAKNKNHAEQTRRAMTLKSLLRALVNMPPHYRYL 320
Query: 288 -----VNWVAWFPFFLFDTDWMGREVYGG-------VVGEKAYDDGVRAGSLGLMINAVV 335
+ W A+ LF TD+MG+ VY G Y+ GV G G IN+V
Sbjct: 321 CISHLIGWTAFLSNMLFFTDFMGQIVYRGDPYSAHNSTEFLIYERGVEVGCWGFCINSVF 380
Query: 336 LAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLITKTAEHDRLISGGATVGA-PS 394
+ S + L + G+K L+ + + G +G P+
Sbjct: 381 SSLYSYFQKVLVSYI-GLKGLYFTGYLLFGL-------------------GTGFIGLFPN 420
Query: 395 SGVKAGAIAFFGVMGIPLAINFSVPFALASIY--------SSAAGA-------GQGLSLG 439
+ FGVM L ++VPF L + Y A G G+G+
Sbjct: 421 VYSTLVLCSLFGVMSSTL---YTVPFNLITEYHREEEKERQQAPGGDPDNSVRGKGMDCA 477
Query: 440 VLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLL 489
L + + Q+LV GG G V+ V A+ L+
Sbjct: 478 TLTCMVQLAQILVG----------GGLGFLVNTAGTVVVVVITASAVALI 517
>PAST_RAT (Q8K4S3) Proton-associated sugar transporter A (PAST-A)
Length = 751
Score = 96.7 bits (239), Expect = 1e-19
Identities = 58/213 (27%), Positives = 105/213 (49%), Gaps = 13/213 (6%)
Query: 39 IAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFG 98
I GI+F +A++ + +TP + +G+P S +W P+ G L+QP++G +SDR T RFG
Sbjct: 93 ILFGIEFSYAMETAYVTPVLLQMGLPDQLYSLVWFISPILGFLLQPLLGAWSDRCTSRFG 152
Query: 99 RRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFGFWILDVANNML 158
RRRPFIL A+ + + L+ DIG ++ D + + V G ++D + +
Sbjct: 153 RRRPFILVLAIGALLGLSLLLNGRDIGMALAD--TATNHKWGILLTVCGVVLMDFSADSA 210
Query: 159 QGPCRAFLGDLAAG-DQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTVTDACP 217
P A++ D+ DQ + + + +G GY G G+H + T
Sbjct: 211 DNPSHAYMMDVCGPVDQDR---GLNIHALMAGLGGGFGYVVG---GIH----WDKTSFGR 260
Query: 218 SFCANLKSCFFFSILLLLILSIAALIYVEDTPL 250
+ L+ + F+ + L + ++ L+ + + PL
Sbjct: 261 ALGGQLRVIYIFTAITLSVTTVFTLVSIPERPL 293
Score = 43.1 bits (100), Expect = 0.002
Identities = 49/210 (23%), Positives = 85/210 (40%), Gaps = 39/210 (18%)
Query: 275 LKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGG-------VVGEKAYDDGVRAGSL 327
+ +P+ L + + W+++ LF TD+MG V+ G + Y+ GV G
Sbjct: 522 MPRPVRNLCVNHFLGWLSFEGMLLFYTDFMGEVVFQGDPKAPHASEAYQKYNSGVTMGCW 581
Query: 328 GLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLITKTAEHDRLISGG 387
G+ I A AF S +E L L V+ L+ I + + + L
Sbjct: 582 GMCIYAFSAAFYSAILEKLEECLS-VRTLYFIAYLLFGLGTGLATL-------------- 626
Query: 388 ATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIY--------SSAAGA--GQGLS 437
+ + V +G++ L ++P++L Y SSA G G G+
Sbjct: 627 ----SRNLYVVLSLCTHYGILFSTLC---TLPYSLLCDYYQSKKFAGSSADGTRRGMGVD 679
Query: 438 LGVLNLAIVVPQMLVSTLSGPWDALFGGGN 467
+ +L+ + Q+LVS + GP + G N
Sbjct: 680 ISLLSCQYFLAQILVSLVLGPLTSAVGSAN 709
>PAST_MOUSE (Q8BIV7) Proton-associated sugar transporter A (PAST-A)
(Deleted in neuroblastoma 5 protein homolog) (DNb-5
homolog)
Length = 751
Score = 95.5 bits (236), Expect = 3e-19
Identities = 59/213 (27%), Positives = 104/213 (48%), Gaps = 13/213 (6%)
Query: 39 IAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFG 98
I GI+F +A++ + +TP + +G+P S +W P+ G L+QP++G +SDR T RFG
Sbjct: 93 ILFGIEFSYAMETAYVTPVLLQMGLPDQLYSLVWFISPILGFLLQPLLGAWSDRCTSRFG 152
Query: 99 RRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAIFVFGFWILDVANNML 158
RRRPFIL A+ + + L+ DIG ++ D + + V G ++D + +
Sbjct: 153 RRRPFILVLAIGALLGLSLLLNGRDIGMALAD--TATNHKWGILLTVCGVVLMDFSADSA 210
Query: 159 QGPCRAFLGDLAAG-DQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTVTDACP 217
P A + D+ DQ + + + +G GY G G+H + T
Sbjct: 211 DNPSHAXMMDVCGPVDQDR---GLNIHALMAGLGGGFGYVVG---GIH----WDKTSFGR 260
Query: 218 SFCANLKSCFFFSILLLLILSIAALIYVEDTPL 250
+ L+ + F+ + L + ++ LI + + PL
Sbjct: 261 ALGGQLRVIYVFTAITLSVTTVLTLISIPERPL 293
Score = 42.0 bits (97), Expect = 0.004
Identities = 50/190 (26%), Positives = 80/190 (41%), Gaps = 25/190 (13%)
Query: 288 VNWVAWFPFFLFDTDWMGREVYGG-------VVGEKAYDDGVRAGSLGLMINAVVLAFMS 340
+ W+++ LF TD+MG V+ G + Y+ GV G G+ I A AF S
Sbjct: 535 LGWLSFEGMLLFYTDFMGEVVFQGDPKAPHTSEAYQKYNSGVTMGCWGMCIYAFSAAFYS 594
Query: 341 LAVEPLGRILGGVKNLWGIVNFILAICMAM-TVLITKTAEHDRLISGGATVGAPSSGVKA 399
+E L L V+ L+ FI + + T L T + ++S T G
Sbjct: 595 AILEKLEECLS-VRTLY----FIAYLAFGLGTGLATLSRNLYVVLSLCTTYG-------- 641
Query: 400 GAIAFFGVMGIPLAINFSVPFALASIYSSAAGA--GQGLSLGVLNLAIVVPQMLVSTLSG 457
I F + +P ++ + SSA G G G+ + +L+ + Q+LVS + G
Sbjct: 642 --ILFSTLCTLPYSLLCDYYQSKKFAGSSADGTRRGMGVDISLLSCQYFLAQILVSLVLG 699
Query: 458 PWDALFGGGN 467
P + G N
Sbjct: 700 PLTSAVGSAN 709
>YD74_SYNY3 (P74168) Hypothetical symporter sll1374
Length = 544
Score = 46.6 bits (109), Expect = 2e-04
Identities = 29/94 (30%), Positives = 46/94 (48%), Gaps = 6/94 (6%)
Query: 26 QSSPLRKMIAVASIAAGI-QFGWALQLSLLTPYV-----QTLGVPHIWASFIWLCGPVSG 79
QS K+ +A G FG A+ ++L Y+ G+P A + + G +
Sbjct: 3 QSLSAEKLHFTTKLAYGAGDFGPAITANILVFYLLFFLTDVAGIPAALAGSVLMIGKIFD 62
Query: 80 LLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAI 113
+ PI+G SDRT R+GRR P++L G + A+
Sbjct: 63 AINDPIIGLLSDRTRSRWGRRLPWMLGGMIPFAL 96
>PAST_HUMAN (Q9Y2W3) Proton-associated sugar transporter A (PAST-A)
(Deleted in neuroblastoma 5 protein) (DNb-5) (Fragment)
Length = 509
Score = 45.1 bits (105), Expect = 4e-04
Identities = 63/258 (24%), Positives = 103/258 (39%), Gaps = 43/258 (16%)
Query: 259 DSPVSCFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGG------- 311
+ P+S G L + K + L + + W+++ LF TD+MG V+ G
Sbjct: 265 EQPLSV-GRLCSTICNMPKALRTLCVNHFLGWLSFEGMLLFYTDFMGEVVFQGDPKAPHT 323
Query: 312 VVGEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMT 371
+ Y+ GV G G+ I A AF S +E L L V+ L+ I
Sbjct: 324 SEAYQKYNSGVTMGCWGMCIYAFSAAFYSAILEKLEEFLS-VRTLYFIAYLAFG------ 376
Query: 372 VLITKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIY----- 426
L +G AT+ V + I + GI + ++P++L Y
Sbjct: 377 -----------LGTGLATLSRNLYVVLSLCITY----GILFSTLCTLPYSLLCDYYQSKK 421
Query: 427 ---SSAAGA--GQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAF---VVGAVMA 478
SSA G G G+ + +L+ + Q+LVS + GP + G N + +V +
Sbjct: 422 FAGSSADGTRRGMGVDISLLSCQYFLAQILVSLVLGPLTSAVGSANGVMYFSSLVSFLGC 481
Query: 479 AVSAIMAIVLLPTPKPAD 496
S++ I +P AD
Sbjct: 482 LYSSLFVIYEIPPSDAAD 499
>YHJ1_SCHPO (Q9C0V8) Probable transporter PB10D8.01
Length = 552
Score = 40.0 bits (92), Expect = 0.014
Identities = 49/168 (29%), Positives = 82/168 (48%), Gaps = 29/168 (17%)
Query: 336 LAFMSLAVEPLGRILGGVKNL-WGIVNFILAICMAMTVLITKTAEHDRL-----ISGGAT 389
L F SL V L L GV + + V+ I+A M M+V+ ++A H + I+GG
Sbjct: 339 LLFTSLNVSDLNSTLVGVASAGYNCVSAIIATYM-MSVIKNQSAFHGAIWYIPSIAGGIA 397
Query: 390 VGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAGAG--QGLSLGVLNLA--- 444
A S K G +A I +A NF +PF ++ +++A+ AG + L+ GV+ +
Sbjct: 398 CVAMSWDHKIGELA-----AIIIASNFGIPFIISLGWTTASCAGYTKKLTRGVMFMIGYA 452
Query: 445 ---IVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMA-AVSAIMAIVL 488
I+ PQM W + PA++V V+A +VS + +++
Sbjct: 453 IANIIGPQM--------WQSRDAPRYYPAWIVQIVIAWSVSPFVLLII 492
>RSG1_HUMAN (P20936) Ras GTPase-activating protein 1
(GTPase-activating protein) (GAP) (Ras p21 protein
activator) (p120GAP) (RasGAP)
Length = 1047
Score = 38.9 bits (89), Expect = 0.032
Identities = 40/129 (31%), Positives = 54/129 (41%), Gaps = 11/129 (8%)
Query: 386 GGATVGAPSSGVKAGAIAFFGV--MGIPLAINFSV---PFALASIYSSAAGAGQGLSLGV 440
G T GA G AG+ A+ V + IP A+ + P + + + G G L
Sbjct: 12 GPVTAGAGGGGAAAGSSAYPAVCRVKIPAALPVAAAPYPGLVETGVAGTLGGGAALGSEF 71
Query: 441 LNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLPTPKPADVAKA 500
L V + + L+G A G A V GA +A S MA+ LPT A
Sbjct: 72 LGAGSVAGALGGAGLTGGGTAA-GVAGAAAGVAGAAVAGPSGDMALTKLPTS-----LLA 125
Query: 501 SSLPPGGGF 509
+L PGGGF
Sbjct: 126 ETLGPGGGF 134
>YNAJ_BACSU (P94488) Hypothetical symporter ynaJ
Length = 463
Score = 38.5 bits (88), Expect = 0.042
Identities = 26/76 (34%), Positives = 36/76 (47%), Gaps = 3/76 (3%)
Query: 53 LLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGA---V 109
LL Y G+ A ++L + L P +G DRT RF R RP++L GA V
Sbjct: 35 LLFFYTDVFGLSAAAAGTMFLVVRIIDALADPFIGTIVDRTNSRFARFRPYLLFGAFPFV 94
Query: 110 AVAISVFLIGYAADIG 125
+AI F +D+G
Sbjct: 95 ILAILCFTTPDFSDMG 110
>YAGG_ECOLI (P75683) Hypothetical symporter yagG
Length = 460
Score = 38.5 bits (88), Expect = 0.042
Identities = 25/80 (31%), Positives = 35/80 (43%), Gaps = 1/80 (1%)
Query: 45 FGWALQLSLLTP-YVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPF 103
F W + LL Y G+ ++L V + P++G DRT R G+ RPF
Sbjct: 21 FVWQATMFLLAYFYTDVFGLSAGIMGTLFLVSRVLDAVTDPLMGLLVDRTRTRHGQFRPF 80
Query: 104 ILAGAVAVAISVFLIGYAAD 123
+L GA+ I L Y D
Sbjct: 81 LLWGAIPFGIVCVLTFYTPD 100
>LACY_LACDE (P22733) Lactose permease (Lactose-proton symporter)
(Lactose transport protein)
Length = 627
Score = 38.5 bits (88), Expect = 0.042
Identities = 34/162 (20%), Positives = 78/162 (47%), Gaps = 18/162 (11%)
Query: 77 VSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKT 136
+ +L+ P++G DRT R+G+ +P+++ G + +++ L+ D G ++
Sbjct: 58 IGEVLLDPLIGNAIDRTESRWGKFKPWVVGGGIISSLA--LLALFTDFGG------INQS 109
Query: 137 RPRAVAIFVFG--FWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVL 194
+P V + +FG + I+D+ + A + L+ +++ +T S F VG+ +
Sbjct: 110 KP-VVYLVIFGIVYLIMDIFYSFKDTGFWAMIPALSLDSREREKT-----STFARVGSTI 163
Query: 195 GYAAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLI 236
G I F+ + A P+ + + FFF++++ ++
Sbjct: 164 GANLVGVVITPIILFFSASKANPN--GDKQGWFFFALIVAIV 203
>YJMB_BACSU (O34961) Hypothetical symporter yjmB
Length = 459
Score = 37.0 bits (84), Expect = 0.12
Identities = 21/71 (29%), Positives = 36/71 (50%), Gaps = 2/71 (2%)
Query: 50 QLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSD--RTTCRFGRRRPFILAG 107
Q+ LL + G+P A I+L + + PIVG D + + G+ RP++L G
Sbjct: 41 QIYLLKYFTDVAGIPAAMAGGIFLVSKLFAAITDPIVGSSIDYRKNIGKRGKFRPYLLIG 100
Query: 108 AVAVAISVFLI 118
++ +A+ LI
Sbjct: 101 SIVLAVLTVLI 111
>YICJ_ECOLI (P31435) Hypothetical symporter yicJ
Length = 460
Score = 37.0 bits (84), Expect = 0.12
Identities = 20/73 (27%), Positives = 38/73 (51%), Gaps = 1/73 (1%)
Query: 49 LQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGA 108
+ L ++ Y G+P + ++L + P +G +DRT R+G+ RP++L GA
Sbjct: 28 VMLYMMFFYTDIFGIPAGFVGTMFLVARALDAISDPCMGLLADRTRSRWGKFRPWVLFGA 87
Query: 109 VAVAISVFLIGYA 121
+ I V ++ Y+
Sbjct: 88 LPFGI-VCVLAYS 99
>ELS_MOUSE (P54320) Elastin precursor (Tropoelastin)
Length = 860
Score = 35.8 bits (81), Expect = 0.27
Identities = 33/97 (34%), Positives = 44/97 (45%), Gaps = 7/97 (7%)
Query: 390 VGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAA-GAGQGLSLGVLNLAIVVP 448
VG P+ GV AG +GV G + + P A A+ +A GAG +LG L VP
Sbjct: 445 VGIPTYGVGAGGFPGYGV-GAGAGLGGASPAAAAAAAKAAKYGAGGAGALGGL-----VP 498
Query: 449 QMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMA 485
+ L G A+ G G +P A AA +A A
Sbjct: 499 GAVPGALPGAVPAVPGAGGVPGAGTPAAAAAAAAAKA 535
>QOX1_BACSU (P34956) Quinol oxidase polypeptide I (EC 1.10.3.-)
(Quinol oxidase aa3-600, subunit qoxB) (Oxidase
aa(3)-600 subunit 1)
Length = 649
Score = 35.4 bits (80), Expect = 0.35
Identities = 42/201 (20%), Positives = 74/201 (35%), Gaps = 35/201 (17%)
Query: 62 GVPHIWASFIWLCG-PVSGLLVQPIVGYYSDRTTCRFGRRRPF----ILAGAVAVAISVF 116
G+P +WA+ W+ G P +++ P G +S+ F R++ F ++ +A+++ F
Sbjct: 266 GMPMLWANLFWIWGHPEVYIVILPAFGIFSE-IISSFARKQLFGYKAMVGSIIAISVLSF 324
Query: 117 LI--------GYAADIGHSMGDDLAKKTRPRAVAIFVFGF--------------WILDVA 154
L+ G +A + + P V IF + F W L
Sbjct: 325 LVWTHHFFTMGNSASVNSFFSITTMAISIPTGVKIFNWLFTMYKGRISFTTPMLWALAFI 384
Query: 155 NNMLQGPCRAFLGDLAAGDQK--KTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTV 212
N + G + +AA D + T + F + + G V AG K+F +
Sbjct: 385 PNFVIGGVTGVMLAMAAADYQYHNTYFLVSHFHYVLIAGTVFACFAGFIFWYPKMFGHKL 444
Query: 213 TDACPS-----FCANLKSCFF 228
+ F CFF
Sbjct: 445 NERIGKWFFWIFMIGFNICFF 465
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.141 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 59,022,398
Number of Sequences: 164201
Number of extensions: 2486652
Number of successful extensions: 8207
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 8125
Number of HSP's gapped (non-prelim): 80
length of query: 510
length of database: 59,974,054
effective HSP length: 115
effective length of query: 395
effective length of database: 41,090,939
effective search space: 16230920905
effective search space used: 16230920905
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0134.5


