

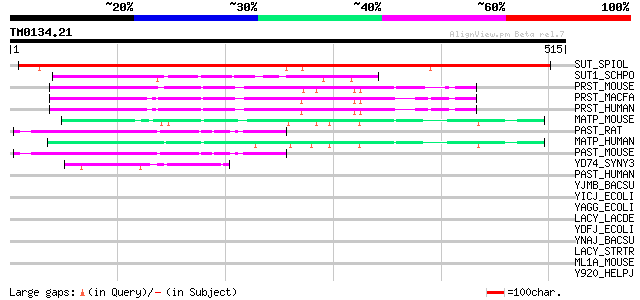
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.21
(515 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SUT_SPIOL (Q03411) Sucrose transport protein (Sucrose permease) ... 638 0.0
SUT1_SCHPO (O14091) General alpha-glucoside permease 143 9e-34
PRST_MOUSE (Q8K0H7) Prostein 110 1e-23
PRST_MACFA (Q95KI5) Prostein (QmoA-10594/QtrA-11310) 108 4e-23
PRST_HUMAN (Q96JT2) Prostein 106 2e-22
MATP_MOUSE (P58355) Membrane-associated transporter protein (AIM... 105 3e-22
PAST_RAT (Q8K4S3) Proton-associated sugar transporter A (PAST-A) 105 4e-22
MATP_HUMAN (Q9UMX9) Membrane-associated transporter protein (AIM... 104 6e-22
PAST_MOUSE (Q8BIV7) Proton-associated sugar transporter A (PAST-... 100 2e-20
YD74_SYNY3 (P74168) Hypothetical symporter sll1374 45 3e-04
PAST_HUMAN (Q9Y2W3) Proton-associated sugar transporter A (PAST-... 41 0.008
YJMB_BACSU (O34961) Hypothetical symporter yjmB 39 0.032
YICJ_ECOLI (P31435) Hypothetical symporter yicJ 37 0.12
YAGG_ECOLI (P75683) Hypothetical symporter yagG 37 0.16
LACY_LACDE (P22733) Lactose permease (Lactose-proton symporter) ... 36 0.21
YDFJ_ECOLI (P77228) Hypothetical metabolite transport protein ydfJ 35 0.47
YNAJ_BACSU (P94488) Hypothetical symporter ynaJ 35 0.61
LACY_STRTR (P23936) Lactose permease (Lactose-proton symporter) ... 35 0.61
ML1A_MOUSE (Q61184) Melatonin receptor type 1A (Mel-1A-R) (Mel1a... 34 0.79
Y920_HELPJ (Q9ZKT1) Hypothetical protein JHP0854 34 1.0
>SUT_SPIOL (Q03411) Sucrose transport protein (Sucrose permease)
(Sucrose-proton symporter)
Length = 525
Score = 638 bits (1646), Expect = 0.0
Identities = 325/511 (63%), Positives = 389/511 (75%), Gaps = 17/511 (3%)
Query: 9 NSTTQNNNSIAPSSFQVE--PAQHAAGPSPLAKMIAVASIAAGVQFGWALQLSLLTPYVQ 66
N NN IA SS +E P + L K+ VAS+AAGVQFGWALQLSLLTPYVQ
Sbjct: 5 NIKNGENNKIAGSSLHLEKNPTTPPEAEATLKKLGLVASVAAGVQFGWALQLSLLTPYVQ 64
Query: 67 LLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIG 126
LLG+PH W+++IWLCGPISG++VQP+VGY SDRCTSRFGRRRPFI AGA VA+AV LIG
Sbjct: 65 LLGIPHTWAAYIWLCGPISGMIVQPLVGYYSDRCTSRFGRRRPFIAAGAALVAVAVGLIG 124
Query: 127 FAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLSTGHHSRIRT 186
FAADIG + GD +PRA+A FV+GFWILDVANN LQGPCRA L D++ G ++ R
Sbjct: 125 FAADIGAASGDPTGNVAKPRAIAVFVVGFWILDVANNTLQGPCRALLADMAAGSQTKTRY 184
Query: 187 ANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTETKACDVFCANLKSCFFFSITLLLVLSG 246
AN FSFFM +GN+ GY AGSY L+ +FPFT+T ACDV+CANLKSCFF SITLL+VL+
Sbjct: 185 ANAFFSFFMALGNIGGYAAGSYSRLYTVFPFTKTAACDVYCANLKSCFFISITLLIVLTI 244
Query: 247 FALFYVHDP--PIGSRREEDDKAPKN---------VFVELFGAFKELKKPMLMLMLVTSL 295
AL V + I +EE+D +N F +L GA K+L KPML+L+LVT+L
Sbjct: 245 LALSVVKERQITIDEIQEEEDLKNRNNSSGCARLPFFGQLIGALKDLPKPMLILLLVTAL 304
Query: 296 NWIAWFPYVLYDTDWMGLEVYGGKLG-SKAYDAGVRAGALGLVLNSVVLGLMSLAVEPLG 354
NWIAWFP++L+DTDWMG EVYGG +G K YD GV AGALGL++NSVVLG+MSL++E L
Sbjct: 305 NWIAWFPFLLFDTDWMGKEVYGGTVGEGKLYDQGVHAGALGLMINSVVLGVMSLSIEGLA 364
Query: 355 RYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDR---RVSGGATIGRPTPGVKAGALMF 411
R +GG KRLW IVNIILAVC+AMT+L+TK AEH R + G A P GVK GAL
Sbjct: 365 RMVGGAKRLWGIVNIILAVCLAMTVLVTKSAEHFRDSHHIMGSAVPPPPPAGVKGGALAI 424
Query: 412 FAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFG 471
FAVLGIPLAIT+S+PFALASI+S+S+G+GQGLSLGVLN+AIV+PQM VS +GPWD++FG
Sbjct: 425 FAVLGIPLAITFSIPFALASIFSASSGSGQGLSLGVLNLAIVVPQMFVSVTSGPWDAMFG 484
Query: 472 GGNLPAFMVGAVMAAVSAVLAMVLLPSPKPE 502
GGNLPAF+VGAV A SAVL+ LLPSP PE
Sbjct: 485 GGNLPAFVVGAVAATASAVLSFTLLPSPPPE 515
>SUT1_SCHPO (O14091) General alpha-glucoside permease
Length = 553
Score = 143 bits (361), Expect = 9e-34
Identities = 96/314 (30%), Positives = 161/314 (50%), Gaps = 29/314 (9%)
Query: 40 MIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDR 99
+IA+ GVQ W+++L TPY+ LG+ +W+S IW+ GP++G+++QPI G SDR
Sbjct: 36 LIALTVSLLGVQLTWSVELGYGTPYLFSLGLRKEWTSIIWIAGPLTGILIQPIAGILSDR 95
Query: 100 CTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSM--GDDLSKKTRPRAVAFFVIGFWI 157
SR GRRRPF+ ++ ++FL+G+A DI + + L K+ + I ++
Sbjct: 96 VNSRIGRRRPFMLCASLLGTFSLFLMGWAPDICLFIFSNEVLMKRV---TIVLATISIYL 152
Query: 158 LDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPF 217
LDVA N++ R+ + D + + AN+ +GVGNVLGYL G Y L++IF F
Sbjct: 153 LDVAVNVVMASTRSLIVD--SVRSDQQHEANSWAGRMIGVGNVLGYLLG-YLPLYRIFSF 209
Query: 218 TETKACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPIGSRREEDDKAPKNVFVELFG 277
VFC SI+L+L ++ +F + RR + K+V E+F
Sbjct: 210 LNFTQLQVFCV------LASISLVLTVTITTIF------VSERRFPPVEHEKSVAGEIFE 257
Query: 278 AFKELKKPMLML-------MLVTSLNWIAWFPYVLYDTDWMGLEV--YGGKLGSKAYDAG 328
F +++ + L V + WFP++ Y T ++G+ + K + +D
Sbjct: 258 FFTTMRQSITALPFTLKRICFVQFFAYFGWFPFLFYITTYVGILYLRHAPKGHEEDWDMA 317
Query: 329 VRAGALGLVLNSVV 342
R G+ L+L +++
Sbjct: 318 TRQGSFALLLFAII 331
>PRST_MOUSE (Q8K0H7) Prostein
Length = 553
Score = 110 bits (274), Expect = 1e-23
Identities = 105/423 (24%), Positives = 184/423 (42%), Gaps = 62/423 (14%)
Query: 38 AKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSS 97
A+++ V + G++ A ++ + P + +GV K+ + + GP+ GLV P++G +S
Sbjct: 16 AQLLLVNLLTFGLEVCLAAGITYVPPLLLEVGVEEKFMTMVLGIGPVLGLVSVPLLGSAS 75
Query: 98 DRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWI 157
D+ R+GRRRPFI+A ++ V +++FLI A + + D TRP +A ++G +
Sbjct: 76 DQWRGRYGRRRPFIWALSLGVLLSLFLIPRAGWLAGLLYPD----TRPLELALLILGVGL 131
Query: 158 LDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPF 217
LD + P A L DL R A ++++F + +G LGYL +
Sbjct: 132 LDFCGQVCFTPLEALLSDLFR-DPDHCRQAFSVYAFMISLGGCLGYLLPAID-------- 182
Query: 218 TETKACDVFCANLKSCFFFSITLL-LVLSGFALFYVHDPPIGSRREEDDKAPKNV----- 271
+T + + C F +TL+ L+ LF + +G + V
Sbjct: 183 WDTSVLAPYLGTQEECLFGLLTLIFLICMAATLFVTEEAVLGPPEPAEGLLVSAVSRRCC 242
Query: 272 -------FVELFGAFKELK-------KPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYG 317
F L F L+ + + L + +W+A + L+ TD++G +Y
Sbjct: 243 PCHVGLAFRNLGTLFPRLQQLCCRMPRTLRRLFVAELCSWMALMTFTLFYTDFVGEGLYQ 302
Query: 318 G----KLGSKA---YDAGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNII 370
G + G++A YD G+R G+LGL L + + SL ++ L + G + ++ +
Sbjct: 303 GVPRAEPGTEARRHYDEGIRMGSLGLFLQCAISLVFSLVMDRLVQKF-GTRSVYLASVMT 361
Query: 371 LAVCMAMTMLITKVAEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALA 430
V A T L V V A A A+ G + +P+ LA
Sbjct: 362 FPVAAAATCLSHSVVV-----------------VTASA----ALTGFTFSALQILPYTLA 400
Query: 431 SIY 433
S+Y
Sbjct: 401 SLY 403
>PRST_MACFA (Q95KI5) Prostein (QmoA-10594/QtrA-11310)
Length = 553
Score = 108 bits (269), Expect = 4e-23
Identities = 106/424 (25%), Positives = 184/424 (43%), Gaps = 64/424 (15%)
Query: 38 AKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSS 97
A+++ + + G++ A ++ + P + +GV K+ + + GP+ GLV P++G +S
Sbjct: 16 AQLLLINLLTFGLEVCLAAGITYVPPLLLEVGVEEKFMTMVLGIGPVLGLVSVPLLGSAS 75
Query: 98 DRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWI 157
D R+GRRRPFI+A ++ + +++FLI A G+ G L RP +A ++G +
Sbjct: 76 DHWRGRYGRRRPFIWALSLGILLSLFLIPRA---GWLAG-LLCPDPRPLELALLILGVGL 131
Query: 158 LDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPF 217
LD + P A L DL R A ++++F + +G LGYL +
Sbjct: 132 LDFCGQVCFTPLEALLSDLFR-DPDHCRQAYSVYAFMISLGGCLGYLLPAID-------- 182
Query: 218 TETKACDVFCANLKSCFFFSITLL-LVLSGFALFYVHDPPIG-SRREEDDKAPK------ 269
+T A + + C F +TL+ L L + +G + E AP
Sbjct: 183 WDTSALAPYLGTQEECLFGLLTLIFLTCVAATLLVAEEAALGPAEPAEGLSAPSLPSHCC 242
Query: 270 ------------NVFVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYG 317
+ L + + + L + +W+A + L+ TD++G +Y
Sbjct: 243 PCWARLAFRNLGALLPRLHQLCCRMPRTLRRLFVAELCSWMALMTFTLFYTDFVGEGLYQ 302
Query: 318 G----KLGSKA---YDAGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNII 370
G +LG++A YD GVR G+LGL L + + SL ++ L + G
Sbjct: 303 GVPRAELGTEARRHYDEGVRMGSLGLFLQCAISLVFSLVMDRLVQRFG------------ 350
Query: 371 LAVCMAMTMLITKVAEHDRRVSGGAT-IGRPTPGVKAGALMFFAVLGIPLAITYSVPFAL 429
+ + VA V+ GAT + V A A A+ G + +P+ L
Sbjct: 351 -----TRAVYLASVAAFP--VAAGATCLSHSVAVVTASA----ALTGFTFSALQILPYTL 399
Query: 430 ASIY 433
AS+Y
Sbjct: 400 ASLY 403
>PRST_HUMAN (Q96JT2) Prostein
Length = 553
Score = 106 bits (264), Expect = 2e-22
Identities = 106/424 (25%), Positives = 183/424 (43%), Gaps = 64/424 (15%)
Query: 38 AKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSS 97
A+++ V + G++ A ++ + P + +GV K+ + + GP+ GLV P++G +S
Sbjct: 16 AQLLLVNLLTFGLEVCLAAGITYVPPLLLEVGVEEKFMTMVLGIGPVLGLVCVPLLGSAS 75
Query: 98 DRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWI 157
D R+GRRRPFI+A ++ + +++FLI A G+ G L RP +A ++G +
Sbjct: 76 DHWRGRYGRRRPFIWALSLGILLSLFLIPRA---GWLAG-LLCPDPRPLELALLILGVGL 131
Query: 158 LDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPF 217
LD + P A L DL R A ++++F + +G LGYL +
Sbjct: 132 LDFCGQVCFTPLEALLSDLFR-DPDHCRQAYSVYAFMISLGGCLGYLLPAID-------- 182
Query: 218 TETKACDVFCANLKSCFFFSITLL-LVLSGFALFYVHDPPIG-SRREEDDKAPK------ 269
+T A + + C F +TL+ L L + +G + E AP
Sbjct: 183 WDTSALAPYLGTQEECLFGLLTLIFLTCVAATLLVAEEAALGPTEPAEGLSAPSLSPHCC 242
Query: 270 ------------NVFVELFGAFKELKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYG 317
+ L + + + L + +W+A + L+ TD++G +Y
Sbjct: 243 PCRARLAFRNLGALLPRLHQLCCRMPRTLRRLFVAELCSWMALMTFTLFYTDFVGEGLYQ 302
Query: 318 G----KLGSKA---YDAGVRAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNII 370
G + G++A YD GVR G+LGL L + + SL ++ L + G
Sbjct: 303 GVPRAEPGTEARRHYDEGVRMGSLGLFLQCAISLVFSLVMDRLVQRFG------------ 350
Query: 371 LAVCMAMTMLITKVAEHDRRVSGGAT-IGRPTPGVKAGALMFFAVLGIPLAITYSVPFAL 429
+ + VA V+ GAT + V A A A+ G + +P+ L
Sbjct: 351 -----TRAVYLASVAAFP--VAAGATCLSHSVAVVTASA----ALTGFTFSALQILPYTL 399
Query: 430 ASIY 433
AS+Y
Sbjct: 400 ASLY 403
>MATP_MOUSE (P58355) Membrane-associated transporter protein (AIM-1
protein) (Melanoma antigen AIM1) (Underwhite protein)
Length = 530
Score = 105 bits (262), Expect = 3e-22
Identities = 117/523 (22%), Positives = 210/523 (39%), Gaps = 124/523 (23%)
Query: 49 GVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRR 108
G +F +A++ + +TP + +G+P S +WL PI G ++QP+VG +SD C +R+GRRR
Sbjct: 44 GREFCYAVEAAYVTPVLLSVGLPKSLYSMVWLLSPILGFLLQPVVGSASDHCRARWGRRR 103
Query: 109 PFIFAGAIAVAIAVFLIGFAADIGYSMGDDL--SKKTRPR-----AVAFFVIGFWILDVA 161
P+I AI + L+G A Y GD + + PR A++ ++G + D +
Sbjct: 104 PYILTLAI-----MMLLGMAL---YLNGDAVVSALVANPRQKLIWAISITMVGVVLFDFS 155
Query: 162 NNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTETK 221
+ + GP +A+L D+ + H + F G G LGY+ G+ +H
Sbjct: 156 ADFIDGPIKAYLFDVCS--HQDKEKGLHYHALFTGFGGALGYILGAIDWVH-------LD 206
Query: 222 ACDVFCANLKSCFFFSITLLLVLSGFALFYVHDPPI-------GSRREEDDKAPKNVFVE 274
+ + FFFS +L++ L + + P+ S+++ + +
Sbjct: 207 LGRLLGTEFQVMFFFSALVLILCFITHLCSIPEAPLRDAATDPPSQQDPQGSSLSASGMH 266
Query: 275 LFGAFKELK-----------------------KPMLMLMLVTSL---------------- 295
+G+ +++K + M M L+ +L
Sbjct: 267 EYGSIEKVKNGGADTEQPVQEWKNKKPSGQSQRTMSMKSLLRALVNMPSHYRCLCVSHLI 326
Query: 296 NWIAWFPYVLYDTDWMGLEVY-GGKLGSK------AYDAGVRAGALGLVLNSVVLGLMSL 348
W A+ +L+ TD+MG VY G G+ Y+ GV G GL +NSV + S
Sbjct: 327 GWTAFLSNMLFFTDFMGQIVYHGDPYGAHNSTEFLIYERGVEVGCWGLCINSVFSSVYSY 386
Query: 349 AVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGATIGRPTPGVKAGA 408
+ + Y+ G+K L+ + ++ + L V
Sbjct: 387 FQKAMVSYI-GLKGLYFMGYLLFGLGTGFIGLFPNV---------------------YST 424
Query: 409 LMFFAVLGIPLAITYSVPFALASIY---------------SSSTGAGQGLSLGVLNVAIV 453
L+ ++ G+ + Y+VPF L + Y + G G+G+ L +
Sbjct: 425 LVLCSMFGVMSSTLYTVPFNLIAEYHREEEKEKGQEAPGGPDNQGRGKGVDCAALTCMVQ 484
Query: 454 IPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLL 496
+ Q++V GG M G+V+ V A+ L+
Sbjct: 485 LAQILVG----------GGLGFLVNMAGSVVVVVITASAVSLI 517
>PAST_RAT (Q8K4S3) Proton-associated sugar transporter A (PAST-A)
Length = 751
Score = 105 bits (261), Expect = 4e-22
Identities = 70/254 (27%), Positives = 121/254 (47%), Gaps = 20/254 (7%)
Query: 4 PPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAGVQFGWALQLSLLTP 63
PPPPPN+ P ++ + +++ I G++F +A++ + +TP
Sbjct: 60 PPPPPNT---------PCPIELVDFEDLHPQRSFWELLFNGCILFGIEFSYAMETAYVTP 110
Query: 64 YVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAVF 123
+ +G+P + S +W PI G ++QP++G SDRCTSRFGRRRPFI AI + +
Sbjct: 111 VLLQMGLPDQLYSLVWFISPILGFLLQPLLGAWSDRCTSRFGRRRPFILVLAIGALLGLS 170
Query: 124 LIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLSTGHHSR 183
L+ DIG ++ D + + V G ++D + + P A++ D+ G +
Sbjct: 171 LLLNGRDIGMALAD--TATNHKWGILLTVCGVVLMDFSADSADNPSHAYMMDV-CGPVDQ 227
Query: 184 IRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTETKACDVFCANLKSCFFFSITLLLV 243
R N I + G+G GY+ GG+H + +T L+ + F+ L V
Sbjct: 228 DRGLN-IHALMAGLGGGFGYVV---GGIH----WDKTSFGRALGGQLRVIYIFTAITLSV 279
Query: 244 LSGFALFYVHDPPI 257
+ F L + + P+
Sbjct: 280 TTVFTLVSIPERPL 293
Score = 40.4 bits (93), Expect = 0.011
Identities = 47/210 (22%), Positives = 86/210 (40%), Gaps = 39/210 (18%)
Query: 282 LKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLGS-------KAYDAGVRAGAL 334
+ +P+ L + L W+++ +L+ TD+MG V+ G + + Y++GV G
Sbjct: 522 MPRPVRNLCVNHFLGWLSFEGMLLFYTDFMGEVVFQGDPKAPHASEAYQKYNSGVTMGCW 581
Query: 335 GLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGG 394
G+ + + S +E L L V+ L+ I ++ + +G
Sbjct: 582 GMCIYAFSAAFYSAILEKLEECLS-VRTLYFIAYLLFGLG-----------------TGL 623
Query: 395 ATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSS----------TGAGQGLS 444
AT+ R V L GI + ++P++L Y S T G G+
Sbjct: 624 ATLSRNLYVV----LSLCTHYGILFSTLCTLPYSLLCDYYQSKKFAGSSADGTRRGMGVD 679
Query: 445 LGVLNVAIVIPQMIVSALNGPWDSLFGGGN 474
+ +L+ + Q++VS + GP S G N
Sbjct: 680 ISLLSCQYFLAQILVSLVLGPLTSAVGSAN 709
>MATP_HUMAN (Q9UMX9) Membrane-associated transporter protein (AIM-1
protein) (Melanoma antigen AIM1)
Length = 530
Score = 104 bits (259), Expect = 6e-22
Identities = 118/522 (22%), Positives = 206/522 (38%), Gaps = 96/522 (18%)
Query: 36 PLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGY 95
P +++I + G +F +A++ + +TP + +G+P S +W PI G ++QP+VG
Sbjct: 31 PTSRLIMHSMAMFGREFCYAVEAAYVTPVLLSVGLPSSLYSIVWFLSPILGFLLQPVVGS 90
Query: 96 SSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGF 155
+SD C SR+GRRRP+I + + + + L A + ++ + +K A++ +IG
Sbjct: 91 ASDHCRSRWGRRRPYILTLGVMMLVGMALYLNGATVVAALIANPRRKL-VWAISVTMIGV 149
Query: 156 WILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLH-KI 214
+ D A + + GP +A+L D+ + H + F G G LGYL G+ H ++
Sbjct: 150 VLFDFAADFIDGPIKAYLFDVCS--HQDKEKGLHYHALFTGFGGALGYLLGAIDWAHLEL 207
Query: 215 FPFTETKACDVF---------CANLKSCFFFSITLLLVLSGFALFYV-HDPPIGS----- 259
T+ +F C + C L V G DPP+ S
Sbjct: 208 GRLLGTEFQVMFFFSALVLTLCFTVHLCSISEAPLTEVAKGIPPQQTPQDPPLSSDGMYE 267
Query: 260 --RREEDDKAPKNVFVELFGA-----FKELKKPMLMLMLVTSL----------------N 296
E+ N + + GA ++ ++ M + L+ +L
Sbjct: 268 YGSIEKVKNGYVNPELAMQGAKNKNHAEQTRRAMTLKSLLRALVNMPPHYRYLCISHLIG 327
Query: 297 WIAWFPYVLYDTDWMGLEVYGGKLGSK-------AYDAGVRAGALGLVLNSVVLGLMSLA 349
W A+ +L+ TD+MG VY G S Y+ GV G G +NSV L S
Sbjct: 328 WTAFLSNMLFFTDFMGQIVYRGDPYSAHNSTEFLIYERGVEVGCWGFCINSVFSSLYSYF 387
Query: 350 VEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGATIGRPTPGVKAGAL 409
+ L Y+ G+K L+ ++ + L V L
Sbjct: 388 QKVLVSYI-GLKGLYFTGYLLFGLGTGFIGLFPNV---------------------YSTL 425
Query: 410 MFFAVLGIPLAITYSVPFALASIY---------------SSSTGAGQGLSLGVLNVAIVI 454
+ ++ G+ + Y+VPF L + Y ++ G+G+ L + +
Sbjct: 426 VLCSLFGVMSSTLYTVPFNLITEYHREEEKERQQAPGGDPDNSVRGKGMDCATLTCMVQL 485
Query: 455 PQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVLAMVLL 496
Q++V GG G V+ V A+ L+
Sbjct: 486 AQILVG----------GGLGFLVNTAGTVVVVVITASAVALI 517
>PAST_MOUSE (Q8BIV7) Proton-associated sugar transporter A (PAST-A)
(Deleted in neuroblastoma 5 protein homolog) (DNb-5
homolog)
Length = 751
Score = 99.8 bits (247), Expect = 2e-20
Identities = 69/254 (27%), Positives = 118/254 (46%), Gaps = 20/254 (7%)
Query: 4 PPPPPNSTTQNNNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAGVQFGWALQLSLLTP 63
PPPPPN+ P ++ +++ I G++F +A++ + +TP
Sbjct: 60 PPPPPNT---------PCPIELVDFGDLHPQRSFWELLFNGCILFGIEFSYAMETAYVTP 110
Query: 64 YVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAVF 123
+ +G+P + S +W PI G ++QP++G SDRCTSRFGRRRPFI AI + +
Sbjct: 111 VLLQMGLPDQLYSLVWFISPILGFLLQPLLGAWSDRCTSRFGRRRPFILVLAIGALLGLS 170
Query: 124 LIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLSTGHHSR 183
L+ DIG ++ D + + V G ++D + + P A + D+ G +
Sbjct: 171 LLLNGRDIGMALAD--TATNHKWGILLTVCGVVLMDFSADSADNPSHAXMMDV-CGPVDQ 227
Query: 184 IRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTETKACDVFCANLKSCFFFSITLLLV 243
R N I + G+G GY+ GG+H + +T L+ + F+ L V
Sbjct: 228 DRGLN-IHALMAGLGGGFGYVV---GGIH----WDKTSFGRALGGQLRVIYVFTAITLSV 279
Query: 244 LSGFALFYVHDPPI 257
+ L + + P+
Sbjct: 280 TTVLTLISIPERPL 293
Score = 38.9 bits (89), Expect = 0.032
Identities = 45/197 (22%), Positives = 79/197 (39%), Gaps = 39/197 (19%)
Query: 295 LNWIAWFPYVLYDTDWMGLEVYGGKLGS-------KAYDAGVRAGALGLVLNSVVLGLMS 347
L W+++ +L+ TD+MG V+ G + + Y++GV G G+ + + S
Sbjct: 535 LGWLSFEGMLLFYTDFMGEVVFQGDPKAPHTSEAYQKYNSGVTMGCWGMCIYAFSAAFYS 594
Query: 348 LAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGGATIGRPTPGVKAG 407
+E L L V+ L+ I + + +G AT+ R V
Sbjct: 595 AILEKLEECLS-VRTLYFIAYLAFGLG-----------------TGLATLSRNLYVV--- 633
Query: 408 ALMFFAVLGIPLAITYSVPFALASIYSSS----------TGAGQGLSLGVLNVAIVIPQM 457
L GI + ++P++L Y S T G G+ + +L+ + Q+
Sbjct: 634 -LSLCTTYGILFSTLCTLPYSLLCDYYQSKKFAGSSADGTRRGMGVDISLLSCQYFLAQI 692
Query: 458 IVSALNGPWDSLFGGGN 474
+VS + GP S G N
Sbjct: 693 LVSLVLGPLTSAVGSAN 709
>YD74_SYNY3 (P74168) Hypothetical symporter sll1374
Length = 544
Score = 45.4 bits (106), Expect = 3e-04
Identities = 41/161 (25%), Positives = 75/161 (46%), Gaps = 17/161 (10%)
Query: 52 FGWALQLSLLTPYV-----QLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGR 106
FG A+ ++L Y+ + G+P + + + G I + PI+G SDR SR+GR
Sbjct: 23 FGPAITANILVFYLLFFLTDVAGIPAALAGSVLMIGKIFDAINDPIIGLLSDRTRSRWGR 82
Query: 107 RRPFIFAGAIAVAI---AVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVANN 163
R P++ G I A+ A +LI +D D L+ + ++V ++
Sbjct: 83 RLPWMLGGMIPFALFYTAQWLIPHFSD------DRLTNQW--GLFIYYVAIAMAFNLCYT 134
Query: 164 MLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYL 204
+ P A +L+ ++ R R + F+F +G G++L +
Sbjct: 135 TVNLPYTALTPELTQNYNERTRLNSFRFAFSIG-GSILSLI 174
>PAST_HUMAN (Q9Y2W3) Proton-associated sugar transporter A (PAST-A)
(Deleted in neuroblastoma 5 protein) (DNb-5) (Fragment)
Length = 509
Score = 40.8 bits (94), Expect = 0.008
Identities = 46/210 (21%), Positives = 87/210 (40%), Gaps = 39/210 (18%)
Query: 282 LKKPMLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLGS-------KAYDAGVRAGAL 334
+ K + L + L W+++ +L+ TD+MG V+ G + + Y++GV G
Sbjct: 280 MPKALRTLCVNHFLGWLSFEGMLLFYTDFMGEVVFQGDPKAPHTSEAYQKYNSGVTMGCW 339
Query: 335 GLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDRRVSGG 394
G+ + + S +E L +L V+ L+ I + + +G
Sbjct: 340 GMCIYAFSAAFYSAILEKLEEFLS-VRTLYFIAYLAFGLG-----------------TGL 381
Query: 395 ATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSS----------TGAGQGLS 444
AT+ R V + + + GI + ++P++L Y S T G G+
Sbjct: 382 ATLSRNLYVVLSLCITY----GILFSTLCTLPYSLLCDYYQSKKFAGSSADGTRRGMGVD 437
Query: 445 LGVLNVAIVIPQMIVSALNGPWDSLFGGGN 474
+ +L+ + Q++VS + GP S G N
Sbjct: 438 ISLLSCQYFLAQILVSLVLGPLTSAVGSAN 467
>YJMB_BACSU (O34961) Hypothetical symporter yjmB
Length = 459
Score = 38.9 bits (89), Expect = 0.032
Identities = 21/77 (27%), Positives = 40/77 (51%), Gaps = 2/77 (2%)
Query: 57 QLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSD--RCTSRFGRRRPFIFAG 114
Q+ LL + + G+P + I+L + + PIVG S D + + G+ RP++ G
Sbjct: 41 QIYLLKYFTDVAGIPAAMAGGIFLVSKLFAAITDPIVGSSIDYRKNIGKRGKFRPYLLIG 100
Query: 115 AIAVAIAVFLIGFAADI 131
+I +A+ LI + ++
Sbjct: 101 SIVLAVLTVLIFLSPNV 117
>YICJ_ECOLI (P31435) Hypothetical symporter yicJ
Length = 460
Score = 37.0 bits (84), Expect = 0.12
Identities = 19/76 (25%), Positives = 36/76 (47%)
Query: 56 LQLSLLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGA 115
+ L ++ Y + G+P + ++L + P +G +DR SR+G+ RP++ GA
Sbjct: 28 VMLYMMFFYTDIFGIPAGFVGTMFLVARALDAISDPCMGLLADRTRSRWGKFRPWVLFGA 87
Query: 116 IAVAIAVFLIGFAADI 131
+ I L D+
Sbjct: 88 LPFGIVCVLAYSTPDL 103
>YAGG_ECOLI (P75683) Hypothetical symporter yagG
Length = 460
Score = 36.6 bits (83), Expect = 0.16
Identities = 23/80 (28%), Positives = 35/80 (43%), Gaps = 1/80 (1%)
Query: 52 FGWALQLSLLTP-YVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPF 110
F W + LL Y + G+ ++L + V P++G DR +R G+ RPF
Sbjct: 21 FVWQATMFLLAYFYTDVFGLSAGIMGTLFLVSRVLDAVTDPLMGLLVDRTRTRHGQFRPF 80
Query: 111 IFAGAIAVAIAVFLIGFAAD 130
+ GAI I L + D
Sbjct: 81 LLWGAIPFGIVCVLTFYTPD 100
>LACY_LACDE (P22733) Lactose permease (Lactose-proton symporter)
(Lactose transport protein)
Length = 627
Score = 36.2 bits (82), Expect = 0.21
Identities = 32/119 (26%), Positives = 56/119 (46%), Gaps = 12/119 (10%)
Query: 84 ISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYSMGDDLSKKT 143
I +++ P++G + DR SR+G+ +P++ G I ++A L+ D G G + SK
Sbjct: 58 IGEVLLDPLIGNAIDRTESRWGKFKPWVVGGGIISSLA--LLALFTDFG---GINQSKPV 112
Query: 144 RPRAVAFFVIGFWILDVANNMLQGPCRAFLGDLSTGHHSRIRTANTIFSFFMGVGNVLG 202
+ F I + I+D+ + A + LS R +T S F VG+ +G
Sbjct: 113 --VYLVIFGIVYLIMDIFYSFKDTGFWAMIPALSLDSREREKT-----STFARVGSTIG 164
>YDFJ_ECOLI (P77228) Hypothetical metabolite transport protein ydfJ
Length = 427
Score = 35.0 bits (79), Expect = 0.47
Identities = 30/113 (26%), Positives = 48/113 (41%), Gaps = 11/113 (9%)
Query: 29 QHAAGPSPLAKMIAVAS--IAAGVQFGWALQLSLLTPY-----VQLLGVPHKWSSFIWLC 81
Q A P+P M S +A G++FG A L+ + VQ L + +
Sbjct: 198 QPTAKPAPAGSMFQSKSFWLATGLRFGQAGNSGLIQTFLAGYLVQTLLFNKAIPTDALMI 257
Query: 82 GPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAVFLIGFAADIGYS 134
I G + P +G+ SD+ GRR P+I A+ +A ++ D Y+
Sbjct: 258 SSILGFMTIPFLGWLSDK----IGRRIPYIIMNTSAIVLAWPMLSIIVDKSYA 306
>YNAJ_BACSU (P94488) Hypothetical symporter ynaJ
Length = 463
Score = 34.7 bits (78), Expect = 0.61
Identities = 23/76 (30%), Positives = 36/76 (47%), Gaps = 3/76 (3%)
Query: 60 LLTPYVQLLGVPHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGA---I 116
LL Y + G+ + ++L I + P +G DR SRF R RP++ GA +
Sbjct: 35 LLFFYTDVFGLSAAAAGTMFLVVRIIDALADPFIGTIVDRTNSRFARFRPYLLFGAFPFV 94
Query: 117 AVAIAVFLIGFAADIG 132
+AI F +D+G
Sbjct: 95 ILAILCFTTPDFSDMG 110
>LACY_STRTR (P23936) Lactose permease (Lactose-proton symporter)
(Lactose transport protein)
Length = 634
Score = 34.7 bits (78), Expect = 0.61
Identities = 67/303 (22%), Positives = 127/303 (41%), Gaps = 38/303 (12%)
Query: 58 LSLLTPYVQLLGVPHKWSSFIWLCGPISGLV------VQPIVGYSSDRCTSRFGRRRPFI 111
+ +T ++ G P + S ++ L I ++ + P++G D +++G+ +P++
Sbjct: 33 IMFVTTHLFNTGDPKQNSHYVLLITNIISILRILEVFIDPLIGNMIDNTNTKYGKFKPWV 92
Query: 112 FAGAIAVAIAVFLIGFAADIGYSMGDDLSKKTRPRAVAFFVIGFWILDVANNMLQGPCRA 171
G I +I + L+ D+G L+K + F I + ++DV ++ +
Sbjct: 93 VGGGIISSITLLLL--FTDLG-----GLNKTNPFLYLVLFGIIYLVMDVFYSIKDIGFWS 145
Query: 172 FLGDLSTGHHSRIRTANTIFSFFMGVGNVLGYLAGSYGGLHKIFPFTETKACDVFCANLK 231
+ LS H R + A F +G+ +G + + F+ T +
Sbjct: 146 MIPALSLDSHEREKMAT-----FARIGSTIGANIVGVAIMPIVLFFSMTNNSGSGDKSGW 200
Query: 232 SCFFFSITLLLVLSGFALFYVHDPPIGSRREEDDKAPKNVFVELFGAFKELKKPMLMLML 291
F F + L+ V++ A+ IG+R E N L FK L + ++ L
Sbjct: 201 FWFAFIVALIGVITSIAV------GIGTREVESKIRDNNEKTSLKQVFKVLGQNDQLMWL 254
Query: 292 VTSLNWIAWFPYVLYDTDWMGLEVYGGK--LGSKAYDAGVRAGALGLVLNSVVLGLMSLA 349
SL + WF Y L L++Y LG D+G + GL + V+GL+S++
Sbjct: 255 --SLGY--WF-YGLGINTLNALQLYYFTFILG----DSGKYSILYGL---NTVVGLVSVS 302
Query: 350 VEP 352
+ P
Sbjct: 303 LFP 305
>ML1A_MOUSE (Q61184) Melatonin receptor type 1A (Mel-1A-R) (Mel1a
melatonin receptor)
Length = 353
Score = 34.3 bits (77), Expect = 0.79
Identities = 25/108 (23%), Positives = 50/108 (46%), Gaps = 8/108 (7%)
Query: 381 ITKVAEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAG 440
++++ ++ GG GRP P A L F + I + I ++ + S+Y +
Sbjct: 5 VSELLNATQQAPGGGEGGRPRPSWLASTLAFILIFTIVVDILGNL-LVILSVYRNKKLRN 63
Query: 441 QG----LSLGVLNVAIVI---PQMIVSALNGPWDSLFGGGNLPAFMVG 481
G +SL V ++ + + P ++ S LN W+ + + AF++G
Sbjct: 64 SGNIFVVSLAVADLVVAVYPYPLVLTSILNNGWNLGYLHCQVSAFLMG 111
>Y920_HELPJ (Q9ZKT1) Hypothetical protein JHP0854
Length = 230
Score = 33.9 bits (76), Expect = 1.0
Identities = 48/190 (25%), Positives = 69/190 (36%), Gaps = 40/190 (21%)
Query: 273 VELFGAFKELKKP---MLMLMLVTSLNWIAWFPYVLYDTDWMGLEVYGGKLGSKAYDAGV 329
V FG KP + ML TSL+ + P LG AG+
Sbjct: 69 VAFFGLMFSKSKPGLNLFMLFAFTSLSGVTLVPL----------------LGMVIAKAGL 112
Query: 330 RAGALGLVLNSVVLGLMSLAVEPLGRYLGGVKRLWAIVNIILAVCMAMTMLITKVAEHDR 389
A L + ++V GLMS+ L + ++ I I++ VC + +
Sbjct: 113 GAVWQALGMTTIVFGLMSVYALKTKNDLANMGKMLFIALIVVVVCSLINLF--------- 163
Query: 390 RVSGGATIGRPT-PGVKAGALMFFAVLGIPLAITYSVPFALASIYSSSTGAGQGLSLGVL 448
+G P V AGA + + L I Y + +Y S A L L L
Sbjct: 164 -------LGSPMFQVVIAGA----SAILFSLYIAYDTQNIVKGMYDSPIDAAVSLYLDFL 212
Query: 449 NVAIVIPQMI 458
NV I I Q+I
Sbjct: 213 NVFISILQII 222
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,499,658
Number of Sequences: 164201
Number of extensions: 2621668
Number of successful extensions: 12982
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 12890
Number of HSP's gapped (non-prelim): 82
length of query: 515
length of database: 59,974,054
effective HSP length: 115
effective length of query: 400
effective length of database: 41,090,939
effective search space: 16436375600
effective search space used: 16436375600
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0134.21


