

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0127b.3
(823 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
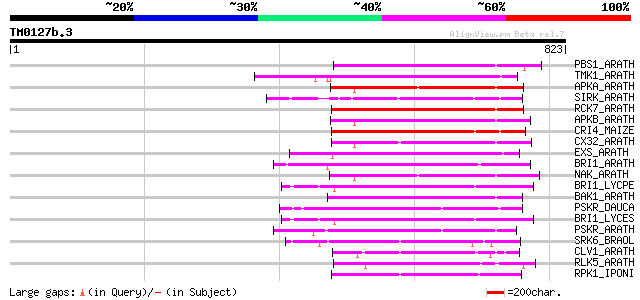
Score E
Sequences producing significant alignments: (bits) Value
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 238 4e-62
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 236 1e-61
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 231 8e-60
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 228 4e-59
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 224 7e-58
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 224 7e-58
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 224 9e-58
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 218 4e-56
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 211 8e-54
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 209 3e-53
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 208 4e-53
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 207 1e-52
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 206 2e-52
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 206 2e-52
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 206 2e-52
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 203 2e-51
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 185 5e-46
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 180 1e-44
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 169 2e-41
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 161 7e-39
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 238 bits (608), Expect = 4e-62
Identities = 135/314 (42%), Positives = 182/314 (56%), Gaps = 8/314 (2%)
Query: 481 ATDNFSEDLVIGSGGFGKVYRGVFKDETKV-AVKRGSCQSQQGLAEFRTEIEMLSQFRHR 539
AT NF D +G GGFG+VY+G +V AVK+ QG EF E+ MLS H
Sbjct: 82 ATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVLMLSLLHHP 141
Query: 540 HLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYL 599
+LV+LIGYC + +R+++YE+M GSL+DHL D L W R++I GAAKGL +L
Sbjct: 142 NLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAGAAKGLEFL 201
Query: 600 HTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEY 659
H +N +I+RD KS+NILLDE K++DFGL+K GP DK +VST V G++GY PEY
Sbjct: 202 HDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRVMGTYGYCAPEY 261
Query: 660 LITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM-RWQERSTIEELVDH 718
+T QLT KSDVYSFGVV E++ GR ID +P + NLV W + +R +L D
Sbjct: 262 AMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFNDRRKFIKLADP 321
Query: 719 HLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEY----ALRLEGVDERSNH 774
L G+ + +L++ + A C+ E RP + DV+ L Y A D R N
Sbjct: 322 RL--KGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYLANQAYDPSKDDSRRNR 379
Query: 775 GGDVSSQIHRSDTG 788
+ I R+D G
Sbjct: 380 DERGARLITRNDDG 393
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 236 bits (603), Expect = 1e-61
Identities = 144/430 (33%), Positives = 225/430 (51%), Gaps = 41/430 (9%)
Query: 363 NVSVGTYSISEASSPEAILNGLEIMKISNSKDSLVFDSEENGKTKVGLIVGLVAGSVVGF 422
NV V T + ++ L+ S S + ++ K +G++ GSV+G
Sbjct: 432 NVVVNTNGNPDIGKDKSSLSSPGSSSPSGGSGSGINGDKDRRGMKSSTFIGIIVGSVLGG 491
Query: 423 FAIVTAFVLLCRRRSRRRNSLHIGDEKGD-------HGATSNYDGTAFFTNSKIG----- 470
+ LL ++R G E + H + N S +
Sbjct: 492 LLSIFLIGLLVFCWYKKRQKRFSGSESSNAVVVHPRHSGSDNESVKITVAGSSVSVGGIS 551
Query: 471 --YRLP--------------------LAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDET 508
Y LP + V++ T+NFS D ++GSGGFG VY+G D T
Sbjct: 552 DTYTLPGTSEVGDNIQMVEAGNMLISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGT 611
Query: 509 KVAVKR--GSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSL 566
K+AVKR + +G AEF++EI +L++ RHRHLV+L+GYC + +E++++YEYM +G+L
Sbjct: 612 KIAVKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTL 671
Query: 567 KDHLF-GSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMA 625
HLF S L WKQRL + + A+G+ YLH ++++ IHRD+K +NILL +++ A
Sbjct: 672 SRHLFEWSEEGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRA 731
Query: 626 KVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGR 685
KVADFGL + P+ K + T + G+FGYL PEY +T ++T K DVYSFGV++ E++ GR
Sbjct: 732 KVADFGLVRLAPE-GKGSIETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGR 790
Query: 686 PVIDPSLPREKMNLVEWIMRW--QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLA 743
+D S P E ++LV W R + ++ ++ +D + + + S+H + A C A
Sbjct: 791 KSLDESQPEESIHLVSWFKRMYINKEASFKKAIDTTIDLDEETLA-SVHTVAELAGHCCA 849
Query: 744 EHGINRPSMG 753
RP MG
Sbjct: 850 REPYQRPDMG 859
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 231 bits (588), Expect = 8e-60
Identities = 128/297 (43%), Positives = 180/297 (60%), Gaps = 14/297 (4%)
Query: 476 AVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETK----------VAVKRGSCQSQQGLAE 525
A ++ AT NF D V+G GGFG V++G +++ +AVK+ + QG E
Sbjct: 59 AELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQGHQE 118
Query: 526 FRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQR 585
+ E+ L QF HRHLV LIGYC E R+++YE+M +GSL++HLF LSWK R
Sbjct: 119 WLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPLSWKLR 178
Query: 586 LEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVS 645
L++ +GAAKGL +LH+ + +I+RD K++NILLD AK++DFGL+K GP DK +VS
Sbjct: 179 LKVALGAAKGLAFLHSSETR-VIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVS 237
Query: 646 TAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMR 705
T V G+ GY PEYL T LT KSDVYSFGVV+ E+L GR +D + P + NLVEW
Sbjct: 238 TRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKP 297
Query: 706 W-QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEY 761
+ + I ++D+ L Q E + + +CL RP+M +V+ HLE+
Sbjct: 298 YLVNKRKIFRVIDNRL--QDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLEH 352
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 228 bits (582), Expect = 4e-59
Identities = 146/381 (38%), Positives = 204/381 (53%), Gaps = 27/381 (7%)
Query: 382 NGLEIMKISNSKDSLVFDSEENGK--TKVGLIVGLVAGSVVGFFAIVTAFVLLCRRRSRR 439
NG ++ + D + DS N K K G I+ LV VVG ++ + L RR ++
Sbjct: 488 NGSLSLRFGRNPDLCLSDSCSNTKKKNKNGYIIPLV---VVGIIVVLLTALALFRRFKKK 544
Query: 440 RNSLHIGDEKGDHGATSNYDGTAFFTNSKIGYRLPLAVIQEATDNFSEDLVIGSGGFGKV 499
+ +G+ G Y + + T+NF VIG GGFGKV
Sbjct: 545 QQRGTLGERNGPLKTAKRY--------------FKYSEVVNITNNFER--VIGKGGFGKV 588
Query: 500 YRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYE 559
Y GV E +VAVK S +S QG EFR E+++L + H +L SL+GYCNE + ++IYE
Sbjct: 589 YHGVINGE-QVAVKVLSEESAQGYKEFRAEVDLLMRVHHTNLTSLVGYCNEINHMVLIYE 647
Query: 560 YMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILL 619
YM +L D+L G + LSW++RL+I + AA+GL YLH G I+HRDVK NILL
Sbjct: 648 YMANENLGDYLAGKRS--FILSWEERLKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILL 705
Query: 620 DENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMF 679
+E L AK+ADFGLS++ +ST V GS GYLDPEY T+Q+ EKSDVYS GVV+
Sbjct: 706 NEKLQAKMADFGLSRSFSVEGSGQISTVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLL 765
Query: 680 EVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQIKSESLHEFVDTAK 739
EV+ G+P I S EK+++ + + I +VD L + S + + A
Sbjct: 766 EVITGQPAI-ASSKTEKVHISDHVRSILANGDIRGIVDQRLRERYDV--GSAWKMSEIAL 822
Query: 740 KCLAEHGINRPSMGDVLWHLE 760
C RP+M V+ L+
Sbjct: 823 ACTEHTSAQRPTMSQVVMELK 843
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 224 bits (571), Expect = 7e-58
Identities = 121/286 (42%), Positives = 174/286 (60%), Gaps = 4/286 (1%)
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVF-KDETKVAVKRGSCQSQQGLAEFRTEIEMLSQF 536
+ EAT NF D +G GGFGKV++G K + VA+K+ QG+ EF E+ LS
Sbjct: 96 LAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVLTLSLA 155
Query: 537 RHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGL 596
H +LV LIG+C E +R+++YEYM +GSL+DHL + L W R++I GAA+GL
Sbjct: 156 DHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKIAAGAARGL 215
Query: 597 HYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLD 656
YLH +I+RD+K +NILL E+ K++DFGL+K GP DK +VST V G++GY
Sbjct: 216 EYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTRVMGTYGYCA 275
Query: 657 PEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM-RWQERSTIEEL 715
P+Y +T QLT KSD+YSFGVV+ E++ GR ID + R+ NLV W +++R ++
Sbjct: 276 PDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFKDRRNFPKM 335
Query: 716 VDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEY 761
VD L GQ L++ + + C+ E RP + DV+ L +
Sbjct: 336 VDPLL--QGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNF 379
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 224 bits (571), Expect = 7e-58
Identities = 126/308 (40%), Positives = 180/308 (57%), Gaps = 14/308 (4%)
Query: 476 AVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETK----------VAVKRGSCQSQQGLAE 525
A ++ AT NF D V+G GGFG V++G ++T +AVK+ + QG E
Sbjct: 60 AELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQGHQE 119
Query: 526 FRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQR 585
+ E+ L QF H +LV LIGYC E R+++YE+M +GSL++HLF + LSW R
Sbjct: 120 WLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLSWTLR 179
Query: 586 LEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVS 645
L++ +GAAKGL +LH ++I+RD K++NILLD AK++DFGL+K GP DK +VS
Sbjct: 180 LKVALGAAKGLAFLHNAET-SVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKSHVS 238
Query: 646 TAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM- 704
T + G++GY PEYL T LT KSDVYS+GVV+ EVL GR +D + P + LVEW
Sbjct: 239 TRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVEWARP 298
Query: 705 RWQERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALR 764
+ + ++D+ L Q E + A +CL RP+M +V+ HLE+
Sbjct: 299 LLANKRKLFRVIDNRL--QDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHLEHIQT 356
Query: 765 LEGVDERS 772
L R+
Sbjct: 357 LNEAGGRN 364
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 224 bits (570), Expect = 9e-58
Identities = 125/291 (42%), Positives = 179/291 (60%), Gaps = 7/291 (2%)
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQS--QQGLAEFRTEIEMLSQ 535
+++AT FSED +G G F V++G+ +D T VAVKR S ++ EF E+++LS+
Sbjct: 498 LEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNELDLLSR 557
Query: 536 FRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATC-LSWKQRLEICIGAAK 594
H HL++L+GYC + SER+++YE+M GSL HL G + + L+W +R+ I + AA+
Sbjct: 558 LNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIAVQAAR 617
Query: 595 GLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGY 654
G+ YLH + +IHRD+KS+NIL+DE+ A+VADFGLS GP +S G+ GY
Sbjct: 618 GIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSELPAGTLGY 677
Query: 655 LDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEE 714
LDPEY LT KSDVYSFGVV+ E+L GR ID + E+ N+VEW + + I
Sbjct: 678 LDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAID--MQFEEGNIVEWAVPLIKAGDIFA 735
Query: 715 LVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRL 765
++D L + E+L + A KC+ G +RPSM V LE+AL L
Sbjct: 736 ILDPVLSPPSDL--EALKKIASVACKCVRMRGKDRPSMDKVTTALEHALAL 784
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 218 bits (556), Expect = 4e-56
Identities = 121/307 (39%), Positives = 182/307 (58%), Gaps = 17/307 (5%)
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDETK----------VAVKRGSCQSQQGLAEFR 527
++ AT NF D ++G GGFGKVYRG T VA+KR + +S QG AE+R
Sbjct: 79 LKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESVQGFAEWR 138
Query: 528 TEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLE 587
+E+ L HR+LV L+GYC E E +++YE+M KGSL+ HLF N W R++
Sbjct: 139 SEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFRRNDP---FPWDLRIK 195
Query: 588 ICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTA 647
I IGAA+GL +LH+ + +I+RD K++NILLD N AK++DFGL+K GP +K +V+T
Sbjct: 196 IVIGAARGLAFLHS-LQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADEKSHVTTR 254
Query: 648 VKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM-RW 706
+ G++GY PEY+ T L KSDV++FGVV+ E++ G + PR + +LV+W+
Sbjct: 255 IMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLVDWLRPEL 314
Query: 707 QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRLE 766
+ +++++D + GQ ++ E C+ NRP M +V+ LE+ L
Sbjct: 315 SNKHRVKQIMDKGI--KGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVLEHIQGLN 372
Query: 767 GVDERSN 773
V RS+
Sbjct: 373 VVPNRSS 379
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 211 bits (536), Expect = 8e-54
Identities = 135/360 (37%), Positives = 194/360 (53%), Gaps = 21/360 (5%)
Query: 415 VAGSVVGFFAIVTAFVLLCRRRSRRRNSLHIGD-EKGDHGATSNYDGTAFFTNSKIGYRL 473
+AG ++GF IV FV RR + + D E+ + + + S R
Sbjct: 830 IAGLMLGFTIIVFVFVFSLRRWAMTKRVKQRDDPERMEESRLKGFVDQNLYFLSGSRSRE 889
Query: 474 PLAV----------------IQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSC 517
PL++ I EATD+FS+ +IG GGFG VY+ E VAVK+ S
Sbjct: 890 PLSINIAMFEQPLLKVRLGDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSE 949
Query: 518 QSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADA 577
QG EF E+E L + +H +LVSL+GYC+ E++++YEYM GSL L
Sbjct: 950 AKTQGNREFMAEMETLGKVKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGML 1009
Query: 578 TCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGP 637
L W +RL+I +GAA+GL +LH G IIHRD+K++NILLD + KVADFGL++
Sbjct: 1010 EVLDWSKRLKIAVGAARGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLIS 1069
Query: 638 DIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPR-EK 696
+ +VST + G+FGY+ PEY + + T K DVYSFGV++ E++ G+ P E
Sbjct: 1070 ACES-HVSTVIAGTFGYIPPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEG 1128
Query: 697 MNLVEWIMRWQERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVL 756
NLV W ++ + +++D L S +K+ L + A CLAE RP+M DVL
Sbjct: 1129 GNLVGWAIQKINQGKAVDVIDPLLV-SVALKNSQL-RLLQIAMLCLAETPAKRPNMLDVL 1186
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 209 bits (531), Expect = 3e-53
Identities = 133/400 (33%), Positives = 206/400 (51%), Gaps = 22/400 (5%)
Query: 391 NSKDSLVFDSEENGKTKVGLIVGLVAGSVVGFFAIVTAFVLLCRRRSRRRNSLHIGDEKG 450
++ D +G+ L G VA ++ F + +L+ R +RR E
Sbjct: 773 SNADGYAHHQRSHGRRPASL-AGSVAMGLLFSFVCIFGLILVGREMRKRRRKKEAELEMY 831
Query: 451 DHGATSNYDGTAFFTNSKIG-----------------YRLPLAVIQEATDNFSEDLVIGS 493
G ++ D TA TN K+ +L A + +AT+ F D +IGS
Sbjct: 832 AEGHGNSGDRTANNTNWKLTGVKEALSINLAAFEKPLRKLTFADLLQATNGFHNDSLIGS 891
Query: 494 GGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSE 553
GGFG VY+ + KD + VA+K+ S QG EF E+E + + +HR+LV L+GYC E
Sbjct: 892 GGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEMETIGKIKHRNLVPLLGYCKVGDE 951
Query: 554 RIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVK 613
R+++YE+M+ GSL+D L L+W R +I IG+A+GL +LH + IIHRD+K
Sbjct: 952 RLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSARGLAFLHHNCSPHIIHRDMK 1011
Query: 614 SANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYS 673
S+N+LLDENL A+V+DFG+++ +D + + G+ GY+ PEY + + + K DVYS
Sbjct: 1012 SSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYS 1071
Query: 674 FGVVMFEVLCG-RPVIDPSLPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQIKSESLH 732
+GVV+ E+L G RP P NLV W+ + + I ++ D L L
Sbjct: 1072 YGVVLLELLTGKRPTDSPDFGDN--NLVGWV-KQHAKLRISDVFDPELMKEDPALEIELL 1128
Query: 733 EFVDTAKKCLAEHGINRPSMGDVLWHLEYALRLEGVDERS 772
+ + A CL + RP+M V+ + G+D +S
Sbjct: 1129 QHLKVAVACLDDRAWRRPTMVQVMAMFKEIQAGSGIDSQS 1168
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 208 bits (530), Expect = 4e-53
Identities = 123/323 (38%), Positives = 187/323 (57%), Gaps = 15/323 (4%)
Query: 475 LAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETK----------VAVKRGSCQSQQGLA 524
L+ ++ AT NF D V+G GGFG V++G + + +AVKR + + QG
Sbjct: 58 LSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQGHR 117
Query: 525 EFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQ 584
E+ EI L Q H +LV LIGYC E+ R+++YE+M +GSL++HLF LSW
Sbjct: 118 EWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPLSWNT 177
Query: 585 RLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYV 644
R+ + +GAA+GL +LH + +I+RD K++NILLD N AK++DFGL++ GP D +V
Sbjct: 178 RVRMALGAARGLAFLHNAQPQ-VIYRDFKASNILLDSNYNAKLSDFGLARDGPMGDNSHV 236
Query: 645 STAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIM 704
ST V G+ GY PEYL T L+ KSDVYSFGVV+ E+L GR ID + P + NLV+W
Sbjct: 237 STRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNLVDWAR 296
Query: 705 RW-QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHL-EYA 762
+ + + ++D L GQ + A C++ +RP+M +++ + E
Sbjct: 297 PYLTNKRRLLRVMDPRL--QGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVKTMEELH 354
Query: 763 LRLEGVDERSNHGGDVSSQIHRS 785
++ E E+ N + + I++S
Sbjct: 355 IQKEASKEQQNPQISIDNIINKS 377
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 207 bits (526), Expect = 1e-52
Identities = 131/386 (33%), Positives = 201/386 (51%), Gaps = 21/386 (5%)
Query: 404 GKTKVGLIVGLVAGSVVGFFAIVTAFVLLCRRRSRRRNSLHIGDEKGDHGATSNYDGTAF 463
G +GL+ L F ++ + +RR ++ +L + H AT+N
Sbjct: 802 GSVAMGLLFSLFC-----IFGLIIVAIETKKRRRKKEAALEAYMDGHSHSATAN--SAWK 854
Query: 464 FTNSKIGYRLPLAVIQ------------EATDNFSEDLVIGSGGFGKVYRGVFKDETKVA 511
FT+++ + LA + EAT+ F D ++GSGGFG VY+ KD + VA
Sbjct: 855 FTSAREALSINLAAFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVA 914
Query: 512 VKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLF 571
+K+ S QG EF E+E + + +HR+LV L+GYC ER+++YEYM+ GSL+D L
Sbjct: 915 IKKLIHVSGQGDREFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLH 974
Query: 572 GSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFG 631
L+W R +I IGAA+GL +LH IIHRD+KS+N+LLDENL A+V+DFG
Sbjct: 975 DRKKTGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFG 1034
Query: 632 LSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPS 691
+++ +D + + G+ GY+ PEY + + + K DVYS+GVV+ E+L G+ D S
Sbjct: 1035 MARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-S 1093
Query: 692 LPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPS 751
NLV W+ + + I ++ D L L + + A CL + RP+
Sbjct: 1094 ADFGDNNLVGWV-KLHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPT 1152
Query: 752 MGDVLWHLEYALRLEGVDERSNHGGD 777
M V+ + G+D S G D
Sbjct: 1153 MIQVMAMFKEIQAGSGMDSTSTIGAD 1178
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 206 bits (525), Expect = 2e-52
Identities = 117/292 (40%), Positives = 173/292 (59%), Gaps = 6/292 (2%)
Query: 472 RLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKR-GSCQSQQGLAEFRTEI 530
R L +Q A+DNFS ++G GGFGKVY+G D T VAVKR ++Q G +F+TE+
Sbjct: 276 RFSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEV 335
Query: 531 EMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICI 590
EM+S HR+L+ L G+C +ER+++Y YM GS+ L L W +R I +
Sbjct: 336 EMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIAL 395
Query: 591 GAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKG 650
G+A+GL YLH + IIHRDVK+ANILLDE A V DFGL+K D +V+TAV+G
Sbjct: 396 GSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKL-MDYKDTHVTTAVRG 454
Query: 651 SFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPS--LPREKMNLVEWIMRWQE 708
+ G++ PEYL T + +EK+DV+ +GV++ E++ G+ D + + + L++W+ +
Sbjct: 455 TIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLK 514
Query: 709 RSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLE 760
+E LVD L G K E + + + A C + RP M +V+ LE
Sbjct: 515 EKKLEALVDVDL--QGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLE 564
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 206 bits (524), Expect = 2e-52
Identities = 128/361 (35%), Positives = 194/361 (53%), Gaps = 8/361 (2%)
Query: 400 SEENGKTKVGLIVGLVAGSVVGFFAIVTAFVLLCRRRSRRRNSLHIGDEKGDHGATSNYD 459
S++N + V + VG G+V F VT ++L R+ R + +
Sbjct: 663 SKKNIRKIVAVAVGTGLGTV--FLLTVTLLIIL---RTTSRGEVDPEKKADADEIELGSR 717
Query: 460 GTAFFTNSKIGYRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQS 519
F N L L I ++T +F++ +IG GGFG VY+ D TKVA+KR S +
Sbjct: 718 SVVLFHNKDSNNELSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDT 777
Query: 520 QQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATC 579
Q EF+ E+E LS+ +H +LV L+GYCN ++++++IY YM+ GSL L
Sbjct: 778 GQMDREFQAEVETLSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPS 837
Query: 580 LSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDI 639
L WK RL I GAA+GL YLH I+HRD+KS+NILL + +A +ADFGL++
Sbjct: 838 LDWKTRLRIARGAAEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPY 897
Query: 640 DKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNL 699
D +V+T + G+ GY+ PEY T K DVYSFGVV+ E+L GR +D PR +L
Sbjct: 898 D-THVTTDLVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDL 956
Query: 700 VEWIMRWQERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHL 759
+ W+++ + E+ D + +E + ++ A +CL E+ RP+ ++ L
Sbjct: 957 ISWVLQMKTEKRESEIFDPFIYDKDH--AEEMLLVLEIACRCLGENPKTRPTTQQLVSWL 1014
Query: 760 E 760
E
Sbjct: 1015 E 1015
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 206 bits (524), Expect = 2e-52
Identities = 131/386 (33%), Positives = 201/386 (51%), Gaps = 21/386 (5%)
Query: 404 GKTKVGLIVGLVAGSVVGFFAIVTAFVLLCRRRSRRRNSLHIGDEKGDHGATSNYDGTAF 463
G +GL+ L F ++ + +RR ++ +L + H AT+N
Sbjct: 802 GSVAMGLLFSLFC-----IFGLIIVAIETKKRRRKKEAALEAYMDGHSHSATAN--SAWK 854
Query: 464 FTNSKIGYRLPLAVIQ------------EATDNFSEDLVIGSGGFGKVYRGVFKDETKVA 511
FT+++ + LA + EAT+ F D ++GSGGFG VY+ KD + VA
Sbjct: 855 FTSAREALSINLAAFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVA 914
Query: 512 VKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLF 571
+K+ S QG EF E+E + + +HR+LV L+GYC ER+++YEYM+ GSL+D L
Sbjct: 915 IKKLIHVSGQGDREFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLH 974
Query: 572 GSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFG 631
L+W R +I IGAA+GL +LH IIHRD+KS+N+LLDENL A+V+DFG
Sbjct: 975 DRKKIGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFG 1034
Query: 632 LSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPS 691
+++ +D + + G+ GY+ PEY + + + K DVYS+GVV+ E+L G+ D S
Sbjct: 1035 MARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-S 1093
Query: 692 LPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPS 751
NLV W+ + + I ++ D L L + + A CL + RP+
Sbjct: 1094 ADFGDNNLVGWV-KLHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPT 1152
Query: 752 MGDVLWHLEYALRLEGVDERSNHGGD 777
M V+ + G+D S G D
Sbjct: 1153 MIQVMAMFKEIQAGSGMDSTSTIGAD 1178
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 203 bits (516), Expect = 2e-51
Identities = 127/363 (34%), Positives = 198/363 (53%), Gaps = 10/363 (2%)
Query: 392 SKDSLVFDSEENGKTKVGLIVGLVAGSVVGFFAIVTAFVLLCRRRSRRRNSLHIGDE--- 448
++ +L+ S + +G+ +G+ GSV +++ VL RRRS + E
Sbjct: 642 TESALIKRSRRSRGGDIGMAIGIAFGSVF-LLTLLSLIVLRARRRSGEVDPEIEESESMN 700
Query: 449 KGDHGATSNYDGTAFFTNSKIGYRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDET 508
+ + G + F +N K L + ++T++F + +IG GGFG VY+ D
Sbjct: 701 RKELGEIGSKLVVLFQSNDK---ELSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGK 757
Query: 509 KVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKD 568
KVA+K+ S Q EF E+E LS+ +H +LV L G+C +++R++IY YME GSL
Sbjct: 758 KVAIKKLSGDCGQIEREFEAEVETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDY 817
Query: 569 HLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVA 628
L N L WK RL I GAAKGL YLH G + I+HRD+KS+NILLDEN + +A
Sbjct: 818 WLHERNDGPALLKWKTRLRIAQGAAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLA 877
Query: 629 DFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVI 688
DFGL++ + +VST + G+ GY+ PEY T K DVYSFGVV+ E+L + +
Sbjct: 878 DFGLARLMSPYE-THVSTDLVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPV 936
Query: 689 DPSLPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGIN 748
D P+ +L+ W+++ + S E+ D + + + + ++ A CL+E+
Sbjct: 937 DMCKPKGCRDLISWVVKMKHESRASEVFDPLI--YSKENDKEMFRVLEIACLCLSENPKQ 994
Query: 749 RPS 751
RP+
Sbjct: 995 RPT 997
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 185 bits (469), Expect = 5e-46
Identities = 118/366 (32%), Positives = 197/366 (53%), Gaps = 23/366 (6%)
Query: 409 GLIVGLVAGSVVGFFAIVTAFVLLCRRRSRRRNS-LHIGDEKGDHGATSN---------Y 458
G I+ L G V ++ F L R++ R + S + I + + + N +
Sbjct: 445 GKIISLTVG--VSVLLLLIMFCLWKRKQKRAKASAISIANTQRNQNLPMNEMVLSSKREF 502
Query: 459 DGTAFFTNSKIGYRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQ 518
G F ++ + + + +AT+NFS +G GGFG VY+G D ++AVKR S
Sbjct: 503 SGEYKFEELELPL-IEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKT 561
Query: 519 SQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADAT 578
S QG EF E+ ++++ +H +LV ++G C E E+++IYEY+E SL +LFG +
Sbjct: 562 SVQGTDEFMNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSK 621
Query: 579 CLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPD 638
L+W +R +I G A+GL YLH S IIHRD+K +NILLD+N++ K++DFG+++
Sbjct: 622 -LNWNERFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFER 680
Query: 639 IDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCG---RPVIDPSLPRE 695
+ + V G++GY+ PEY + +EKSDV+SFGV++ E++ G R + +
Sbjct: 681 DETEANTMKVVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYEND 740
Query: 696 KMNLVEWIMRWQERSTIE----ELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPS 751
++ V W RW+E +E +VD + + + + + C+ E +RP+
Sbjct: 741 LLSYV-W-SRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPA 798
Query: 752 MGDVLW 757
M V+W
Sbjct: 799 MSSVVW 804
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 180 bits (457), Expect = 1e-44
Identities = 110/289 (38%), Positives = 161/289 (55%), Gaps = 21/289 (7%)
Query: 479 QEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKR----GSCQSQQGLAEFRTEIEMLS 534
++ + E+ +IG GG G VYRG + VA+KR G+ +S G F EI+ L
Sbjct: 686 EDVLECLKEENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHG---FTAEIQTLG 742
Query: 535 QFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAK 594
+ RHRH+V L+GY + +++YEYM GSL + L GS L W+ R + + AAK
Sbjct: 743 RIRHRHIVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSKGGH--LQWETRHRVAVEAAK 800
Query: 595 GLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGY 654
GL YLH + I+HRDVKS NILLD + A VADFGL+K D +++ GS+GY
Sbjct: 801 GLCYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGY 860
Query: 655 LDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERST--- 711
+ PEY T ++ EKSDVYSFGVV+ E++ G+ + E +++V W+ +E T
Sbjct: 861 IAPEYAYTLKVDEKSDVYSFGVVLLELIAGKKPVGEF--GEGVDIVRWVRNTEEEITQPS 918
Query: 712 ----IEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVL 756
+ +VD L +G + +H F A C+ E RP+M +V+
Sbjct: 919 DAAIVVAIVDPRL--TGYPLTSVIHVF-KIAMMCVEEEAAARPTMREVV 964
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 169 bits (429), Expect = 2e-41
Identities = 111/318 (34%), Positives = 162/318 (50%), Gaps = 24/318 (7%)
Query: 480 EATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAE----------FRTE 529
E D E VIG G GKVY+ + VAVK+ + + G E F E
Sbjct: 678 EIADCLDEKNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAE 737
Query: 530 IEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEIC 589
+E L RH+ +V L C+ ++++YEYM GSL D L G L W +RL I
Sbjct: 738 VETLGTIRHKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRIA 797
Query: 590 IGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKR--YVSTA 647
+ AA+GL YLH I+HRDVKS+NILLD + AKVADFG++K G + +
Sbjct: 798 LDAAEGLSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSG 857
Query: 648 VKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQ 707
+ GS GY+ PEY+ T ++ EKSD+YSFGVV+ E++ G+ D L + M +W+
Sbjct: 858 IAGSCGYIAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTDSELGDKDM--AKWVCTAL 915
Query: 708 ERSTIEELVDHHLPGSGQIK-SESLHEFVDTAKKCLAEHGINRPSMGDVLWHLE-----Y 761
++ +E ++D L +K E + + + C + +NRPSM V+ L+
Sbjct: 916 DKCGLEPVIDPKL----DLKFKEEISKVIHIGLLCTSPLPLNRPSMRKVVIMLQEVSGAV 971
Query: 762 ALRLEGVDERSNHGGDVS 779
+RS GG +S
Sbjct: 972 PCSSPNTSKRSKTGGKLS 989
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 161 bits (407), Expect = 7e-39
Identities = 98/286 (34%), Positives = 149/286 (51%), Gaps = 6/286 (2%)
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQS-QQGLAEFRTEIEMLSQF 536
+ EAT+N ++ VIG G G +Y+ + AVK+ + G EIE + +
Sbjct: 809 VLEATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGKV 868
Query: 537 RHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGL 596
RHR+L+ L + + +I+Y YME GSL D L +N L W R I +G A GL
Sbjct: 869 RHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKP-LDWSTRHNIAVGTAHGL 927
Query: 597 HYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLD 656
YLH + AI+HRD+K NILLD +L ++DFG++K S V+G+ GY+
Sbjct: 928 AYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMA 987
Query: 657 PEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMR-WQERSTIEEL 715
PE T + +SDVYS+GVV+ E++ + +DPS E ++V W+ W + I+++
Sbjct: 988 PENAFTTVKSRESDVYSYGVVLLELITRKKALDPSFNGE-TDIVGWVRSVWTQTGEIQKI 1046
Query: 716 VDHHLPGS--GQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHL 759
VD L E + E + A +C + RP+M DV+ L
Sbjct: 1047 VDPSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 96,994,995
Number of Sequences: 164201
Number of extensions: 4223754
Number of successful extensions: 14383
Number of sequences better than 10.0: 1631
Number of HSP's better than 10.0 without gapping: 1101
Number of HSP's successfully gapped in prelim test: 530
Number of HSP's that attempted gapping in prelim test: 10601
Number of HSP's gapped (non-prelim): 1923
length of query: 823
length of database: 59,974,054
effective HSP length: 119
effective length of query: 704
effective length of database: 40,434,135
effective search space: 28465631040
effective search space used: 28465631040
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0127b.3


