

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0125.11
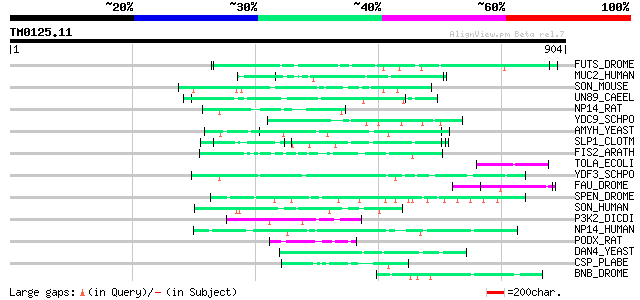
(904 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
FUTS_DROME (Q9W596) Microtubule-associated protein futsch 54 2e-06
MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2) 53 4e-06
SON_MOUSE (Q9QX47) SON protein 52 5e-06
UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89 (Uncoo... 52 9e-06
NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 k... 52 9e-06
YDC9_SCHPO (Q10172) Hypothetical protein C25G10.09c in chromosome I 50 2e-05
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 50 4e-05
SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer... 49 5e-05
FIS2_ARATH (Q9ZNT9) Polycomb group protein FERTILIZATION-INDEPEN... 49 5e-05
TOLA_ECOLI (P19934) TolA protein 48 1e-04
YDF3_SCHPO (Q10475) Probable eukaryotic initiation factor C17C9.03 47 2e-04
FAU_DROME (Q9VGX3) Anoxia up-regulated protein 47 2e-04
SPEN_DROME (Q8SX83) Split ends protein 47 3e-04
SON_HUMAN (P18583) SON protein (SON3) (Negative regulatory eleme... 47 3e-04
P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.13... 47 3e-04
NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130... 47 3e-04
PODX_RAT (Q9WTQ2) Podocalyxin precursor 46 4e-04
DAN4_YEAST (P47179) Cell wall protein DAN4 precursor 46 4e-04
CSP_PLABE (P06915) Circumsporozoite protein precursor (CS) 46 5e-04
BNB_DROME (P29746) Bangles and beads protein 46 5e-04
>FUTS_DROME (Q9W596) Microtubule-associated protein futsch
Length = 5412
Score = 53.9 bits (128), Expect = 2e-06
Identities = 111/563 (19%), Positives = 208/563 (36%), Gaps = 37/563 (6%)
Query: 333 SEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTRVEP 392
S+E+ P ++ + ETS+P+ + DG K+ + V + +
Sbjct: 3213 SQEASRPESEAESLKDAAAPSQETSRPESVTESVKDGKSPVASKEASRPASVAENAKDSA 3272
Query: 393 AAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQP 452
+R E+ P+ S++ + D S + ++ ++ PL S+P
Sbjct: 3273 DESKEQRPESLPQSKAGSIKDEKSPLASKDEAEKSKEESRRESVAEQF--PLVSKEVSRP 3330
Query: 453 AAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPEL 512
A+ S + + S E+PL + ++P + + ES ++ +
Sbjct: 3331 ASVAESVKDEA-----EKSKEESPLMSKEASRPASVAGSVKDEAEKSKEESRRESVAEKS 3385
Query: 513 TIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRK 572
+P + P SV +++ + +S + +P++ + S P++ +ESI+
Sbjct: 3386 PLPSKEASRPASVAESVKDEADK-SKEESRRESGAEKSPLA---SKEASRPASVAESIKD 3441
Query: 573 FMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVD--------PDEPILANPLHEADPLA 624
E KE+ S E P P + RP + + E + + E PLA
Sbjct: 3442 EAEKSKEE-SRRESVAEKSPLPSK-EASRPTSVAESVKDEAEKSKEESRRDSVAEKSPLA 3499
Query: 625 QQAQPDPVQ-QEPVQPDPEQS--VSNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQ 681
+ P E VQ + E+S S SV +PL AS P I
Sbjct: 3500 SKEASRPASVAESVQDEAEKSKEESRRESVAEKSPLASKE-----ASRPASVAESIKDEA 3554
Query: 682 GTSEAHSSNHPTSPETNLSIVPYTSLRPTSLSECINVFNREASLMLRNVQGQTDLSENAE 741
S+ S + ++ L+ + RPTS++E + E S + + S A
Sbjct: 3555 EKSKEESRRESVAEKSPLA--SKEASRPTSVAESVK-DEAEKSKEESSRDSVAEKSPLAS 3611
Query: 742 SVAEKWNSLSTWLVAQVPVMMQHLTAERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEE 801
A + S++ + + + E + A R A + + ++ EE
Sbjct: 3612 KEASRPASVAESVQDEAEKSKEESRRESVAEKSPLASKEASRPASVAESVKDDAEKSKEE 3671
Query: 802 ARR-----KQEQAEEAARQAAAQAEQARLEAERLEAEAEARRLAPVVFTPAASASTPAHQ 856
+RR K A + A + A+ AE + EAE+ + E+ +A P+ AS P
Sbjct: 3672 SRRESVAEKSPLASKEASRPASVAESVKDEAEKSKEESRRESVAEKSPLPSKEASRPTSV 3731
Query: 857 AAQNVPSNSTPPSSSRLELMEQR 879
A SR E + ++
Sbjct: 3732 AESVKDEAEKSKEESRRESVAEK 3754
Score = 45.1 bits (105), Expect = 9e-04
Identities = 111/587 (18%), Positives = 210/587 (34%), Gaps = 81/587 (13%)
Query: 330 EESSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTR 389
E S ES V K +KA+ + E+ KP+ D + +PP K+ + V+ +
Sbjct: 3132 EVSRPESVVGSIKDEKAESRRESVAESVKPESSKDATS----APPSKEHSRPESVLGSLK 3187
Query: 390 VEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPE 449
E RR+ SV DS D S L +S R + + +
Sbjct: 3188 DEGDKTTSRRV----------------SVADSIKDEKS---LLVSQEASRPESEAESLKD 3228
Query: 450 SQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLE 509
+ +Q TS+P S+ S ++P+ + ++P ++ + + E P +L
Sbjct: 3229 AAAPSQETSRPESVTESV---KDGKSPVASKEASRPASVAENAKD-SADESKEQRPESLP 3284
Query: 510 PELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSES 569
++ E P S ++ E + V +S P +VA ES
Sbjct: 3285 QSKAGSIKDEKSPLASKDEAEKSKEESRRESVAEQFPLVSKEVSRPASVA--------ES 3336
Query: 570 IRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQP 629
++ E KE+ + + P G + E + E PL +
Sbjct: 3337 VKDEAEKSKEESPLMSK---EASRPASVAGSVKDEAEKSKEESRRESVAEKSPLPSKEAS 3393
Query: 630 DPVQ-----QEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGAS-----------EPHVQ 673
P ++ E+S + +SP + S P A E +
Sbjct: 3394 RPASVAESVKDEADKSKEESRRESGAEKSPLASKEASRPASVAESIKDEAEKSKEESRRE 3453
Query: 674 TCDIGSPQGTSEAHSSNHPTSPETNLSIVPYTSL---RPTSLSECINVFNREASL---ML 727
+ SP + EA + PTS ++ S R S++E + ++EAS +
Sbjct: 3454 SVAEKSPLPSKEA---SRPTSVAESVKDEAEKSKEESRRDSVAEKSPLASKEASRPASVA 3510
Query: 728 RNVQGQTDLSENA---ESVAEKWNSLSTWLVAQVPVMMQHLTAERDQRIKAAKQRFARRV 784
+VQ + + S+ ESVAEK S A P + + ++ K
Sbjct: 3511 ESVQDEAEKSKEESRRESVAEKSPLASK--EASRPASVAESIKDEAEKSK---------- 3558
Query: 785 ALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQAEQARLEAERLEAEAEARRLAPVVF 844
E+ +R + E + A ++ + A +AE+++ E+ R ++ +
Sbjct: 3559 ---EESRRESVAEKSPLASKEASRPTSVAESVKDEAEKSKEESSRDSVAEKSPLASKEAS 3615
Query: 845 TPAASASTPAHQAAQNVPSNSTPPSSSRLELMEQRLDTHESMLQDMK 891
PA+ A + +A ++ + + + L + S+ + +K
Sbjct: 3616 RPASVAESVQDEAEKSKEESRRESVAEKSPLASKEASRPASVAESVK 3662
Score = 40.4 bits (93), Expect = 0.021
Identities = 87/464 (18%), Positives = 168/464 (35%), Gaps = 57/464 (12%)
Query: 392 PAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQ 451
P + +R P ++R+ + S ++ S D P +LK S+ P +
Sbjct: 2632 PLDEALRTPSAPEHISRADSPAECAS-----EEIASQDKSP--QVLKESSRPAWVAESKD 2684
Query: 452 PAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPE 511
AAQ S LRS + + P + Q PLT+E +
Sbjct: 2685 DAAQLKSSVEDLRSPVASTEISRPASAGETASSPIEEAPKDFAEFEQAEKAVLPLTIELK 2744
Query: 512 LTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIR 571
+P + P ++ P+ S TP S PT VA P+ S+
Sbjct: 2745 GNLPTLSSPVD-VAHGDFPQTS----------------TPTSSPT-VASVQPAELSK--- 2783
Query: 572 KFMEVRKEKVSTLEE---YYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQ 628
+++ K S ++E + CP+ R P E D E + + P ++
Sbjct: 2784 --VDIEKTASSPIDEAPKSLIGCPAEERPESP-AESAKDAAESVEKSKDASRPPSVVEST 2840
Query: 629 PDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSE-----PHLGASEPHVQTCDIGSPQGT 683
+ + P PE S + P V+ S+ P + AS T D+ P
Sbjct: 2841 KADSTKGDISPSPE------SVLEGPKDDVEKSKESSRPPSVSASITGDSTKDVSRPASV 2894
Query: 684 SEAHSSNHPTSPETNLSIVPYTSL------RPTSLSECINVFNREASLMLRNVQGQTDLS 737
E+ H + SI S+ + S + + +++L + + +
Sbjct: 2895 VESVKDEHDKAESRRESIAKVESVIDEAGKSDSKSSSQDSQKDEKSTLASKEASRRESVV 2954
Query: 738 ENAESVAEKWNSLSTWLVAQVPVMMQHLTAERDQRIKAAKQRFARRVAL-----HEQQQR 792
E+++ AEK S ++A + + + D + + V +Q +R
Sbjct: 2955 ESSKDDAEKSESRPESVIASGEPVPRESKSPLDSKDTSRPGSMVESVTAEDEKSEQQSRR 3014
Query: 793 HKLLEAAEEARRKQEQAEEAARQAAAQAEQARLEAERLEAEAEA 836
+ E+ + +K +++EA+R ++ E + + E+ E+ ++
Sbjct: 3015 ESVAESVKADTKKDGKSQEASRPSSVD-ELLKDDDEKQESRRQS 3057
Score = 39.7 bits (91), Expect = 0.037
Identities = 118/616 (19%), Positives = 222/616 (35%), Gaps = 77/616 (12%)
Query: 314 EDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDGGPSP 373
E + ++ + + +E+S + V K AD+ K + E+ ++ P
Sbjct: 3240 ESVTESVKDGKSPVASKEASRPASVAENAKDSADESKEQRPESLPQSKAGSIKDEKSPLA 3299
Query: 374 PKKKKKQVRLVVKPTRVEPAAQ----VVRRIETPPRVTRSSVRSSSKSVVDS-----DAD 424
K + ++ + + +R E A+ V + + P V S + KS +S +A
Sbjct: 3300 SKDEAEKSK---EESRRESVAEQFPLVSKEVSRPASVAESVKDEAEKSKEESPLMSKEAS 3356
Query: 425 LNSFDALPISAMLKRST-----------NPLTLIPESQPAAQTTSQPN----SLRSSFFQ 469
+ A + ++S +PL S+PA+ S + S S +
Sbjct: 3357 RPASVAGSVKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESVKDEADKSKEESRRE 3416
Query: 470 PSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDHSV 529
++PL + ++P ++ + ES ++ + +P + P SV
Sbjct: 3417 SGAEKSPLASKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPTSVAESV 3476
Query: 530 PRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKE---KVSTLEE 586
+E+ + DS + +P++ + S P++ +ES++ E KE + S E+
Sbjct: 3477 KDEAEKSKEESR-RDSVAEKSPLA---SKEASRPASVAESVQDEAEKSKEESRRESVAEK 3532
Query: 587 YYLTCPSPRRYPGPRPERLVDPDEPILANPLHEA----DPLAQQAQPDPVQ-QEPVQPDP 641
L R P E + D E E+ PLA + P E V+ +
Sbjct: 3533 SPLASKEASR-PASVAESIKDEAEKSKEESRRESVAEKSPLASKEASRPTSVAESVKDEA 3591
Query: 642 EQSV--SNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNL 699
E+S S+ SV +PL + VQ EA S + E+
Sbjct: 3592 EKSKEESSRDSVAEKSPLASKEASRPASVAESVQ----------DEAEKSKEESRRESVA 3641
Query: 700 SIVPYTSL---RPTSLSECIN-------------VFNREASLMLRNVQGQTDLSENAESV 743
P S RP S++E + ++ L + ++E+ +
Sbjct: 3642 EKSPLASKEASRPASVAESVKDDAEKSKEESRRESVAEKSPLASKEASRPASVAESVKDE 3701
Query: 744 AEKWNSLSTWLVAQVPVMMQHLTAERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEAR 803
AEK S + A R + + + A + E+ +R + E + A
Sbjct: 3702 AEKSKEESRRESVAEKSPLPSKEASRPTSVAESVKDEAEKSK--EESRRESVAEKSSLAS 3759
Query: 804 RKQEQAEEAARQAAAQAEQARLEAERLEAEAEARRLAPVVFTPAASASTPAHQAAQNVPS 863
+K + A +AE+++ E+ R E+ AE LA + AS PA A
Sbjct: 3760 KKASRPASVAESVKDEAEKSKEESRR-ESVAEKSPLA------SKEASRPASVAESVKDE 3812
Query: 864 NSTPPSSSRLELMEQR 879
SR E + ++
Sbjct: 3813 AEKSKEESRRESVAEK 3828
Score = 36.2 bits (82), Expect = 0.40
Identities = 123/645 (19%), Positives = 230/645 (35%), Gaps = 93/645 (14%)
Query: 310 APAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDG 369
A A E SS I + + E +E++ +PLT + K + P S P D GD
Sbjct: 2709 ASAGETASSPIEEAPKDFAEFEQAEKAVLPLTIELKGNLPT-----LSSPVDVAHGDFPQ 2763
Query: 370 GPSPPKKKKKQVRLVVKPT--RVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNS 427
+P PT V+PA IE + S + + KS++ A+
Sbjct: 2764 TSTPTSS----------PTVASVQPAELSKVDIE---KTASSPIDEAPKSLIGCPAEERP 2810
Query: 428 FDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTG 487
+ + + + + +S+ A++ S S ++ + + +P ++ G
Sbjct: 2811 ------ESPAESAKDAAESVEKSKDASRPPSVVESTKADSTKGDISPSP-----ESVLEG 2859
Query: 488 PSDPTSPITIQYNPESEPLTLEPELTIPVQTEP---EPRMSDHSVPRASERLVSRT---- 540
P D P S ++ + T V E +H + +++
Sbjct: 2860 PKDDVEKSKESSRPPSVSASITGDSTKDVSRPASVVESVKDEHDKAESRRESIAKVESVI 2919
Query: 541 -----TDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTC--PS 593
+D+ SSS + + +A S ++ ES+ + + EK + E + P
Sbjct: 2920 DEAGKSDSKSSSQDSQKDEKSTLA-SKEASRRESVVESSKDDAEKSESRPESVIASGEPV 2978
Query: 594 PRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRS 653
PR P + ++ + E + QQ++ + V E V+ D ++ + + R
Sbjct: 2979 PRESKSPLDSKDTSRPGSMVESVTAEDEKSEQQSRRESVA-ESVKADTKKDGKSQEASRP 3037
Query: 654 PNP---LVDTSEPHLGASEP----HVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVPYTS 706
+ L D E + H +G +A S P+ PE+ + + +
Sbjct: 3038 SSVDELLKDDDEKQESRRQSITGSHKAMSTMGDESPMDKADKSKEPSRPESVAESIKHEN 3097
Query: 707 L---------RPTSLSECIN----------VFNREASLMLRNVQGQTD--LSENAESVAE 745
R S++E I + ++E S V D ESVAE
Sbjct: 3098 TKDEESPLGSRRDSVAESIKSDITKGEKSPLPSKEVSRPESVVGSIKDEKAESRRESVAE 3157
Query: 746 KWNSLSTWLVAQVPVMMQHLTAE------RDQRIKAAKQRFARRVALHEQQQRHKLLEAA 799
S+ P +H E +D+ K +RRV++ + + K L +
Sbjct: 3158 SVKPESSKDATSAPPSKEHSRPESVLGSLKDEGDKTT----SRRVSVADSIKDEKSLLVS 3213
Query: 800 EEARRKQEQAEEAARQAAAQAEQARLEA--------ERLEAEAEARRLAPVVFTPAASAS 851
+EA R + +AE AA E +R E+ + A EA R A V SA
Sbjct: 3214 QEASRPESEAESLKDAAAPSQETSRPESVTESVKDGKSPVASKEASRPASVAENAKDSAD 3273
Query: 852 TPAHQAAQNVPSNSTPPSSSRLELMEQRLDTHESMLQDMKQMMME 896
Q +++P + + + + +S + ++ + E
Sbjct: 3274 ESKEQRPESLPQSKAGSIKDEKSPLASKDEAEKSKEESRRESVAE 3318
Score = 33.1 bits (74), Expect = 3.4
Identities = 119/577 (20%), Positives = 204/577 (34%), Gaps = 88/577 (15%)
Query: 307 KKIAPAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHD--ETSKPDDGND 364
+ +A + +D + K + V+E SDV +K AD+ K E KPD ++
Sbjct: 1738 ESVAESEKDDTDK-----PESVVESVIPASDVVEIEKGAADKEKGVFVSLEIGKPDSPSE 1792
Query: 365 GDNDGGPS----PPKKKKKQVRLVVKPTRVEPAAQVVRR------IETPPRVTRSSVRSS 414
+ GP P+ +++ +V P E + + R ++ V + S R
Sbjct: 1793 VISRPGPVVESVKPESRRESSTEIVLPCHAEDSKEPSRPESKVECLKDESEVLKGSTRRE 1852
Query: 415 SKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQP-----AAQT--TSQPNSLRSSF 467
S + D + + P SA+ ++ P + AAQ+ TS+P S+ S
Sbjct: 1853 SVAESDKSSQPFKETSRPESAVGSMKDESMSKEPSRRESVKDGAAQSRETSRPASVAESA 1912
Query: 468 FQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDH 527
+ + L + + + T S I E PL E E ++
Sbjct: 1913 KDGADD---LKELSRPESTTQSKEAGSI----KDEKSPLASEEASRPASVAESVKDEAEK 1965
Query: 528 SVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEY 587
S + V+ + S P S ++ D + + ES R+ S E+
Sbjct: 1966 SKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRE---------SVAEKS 2016
Query: 588 YLTCPSPRRYPGPRPERLVDPDEPILANPLHEA----DPLAQQAQPDPVQ-QEPVQPDPE 642
L R P E + D E E+ PL + P E ++ + E
Sbjct: 2017 PLPSKEASR-PASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAE 2075
Query: 643 QSV--SNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLS 700
+S S SV +PL P AS P I S+ S + ++ L
Sbjct: 2076 KSKEESRRESVAEKSPL-----PSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLP 2130
Query: 701 IVPYTSLRPTSLSECINVFNREASLMLRNVQGQTDLSENAESVAEKWNSLSTWLVAQVPV 760
+ RP S++E I ++ ++ ESVAEK + +P
Sbjct: 2131 --SKEASRPASVAESI-----------KDEAEKSKEESRRESVAEK---------SPLPS 2168
Query: 761 MMQHLTAERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQA 820
A + IK ++ E+ +R + E + ++ + A +A
Sbjct: 2169 KEASRPASVAESIKDEAEKSK------EESRRESVAEKSPLPSKEASRPASVAESIKDEA 2222
Query: 821 EQARLEAERLEAEAEARRLAPVVFTPAASASTPAHQA 857
E+++ E R E+ AE L P+ AS PA A
Sbjct: 2223 EKSKEETRR-ESVAEKSPL------PSKEASRPASVA 2252
>MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2)
Length = 5179
Score = 52.8 bits (125), Expect = 4e-06
Identities = 64/288 (22%), Positives = 91/288 (31%), Gaps = 36/288 (12%)
Query: 434 SAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTS 493
S + +T PL S P + TT+ P P+ +P T PS PT+
Sbjct: 1439 SPPITTTTTPLPTTTPSPPISTTTTPP---------PTTTPSPPTTTPSPPTTTPSPPTT 1489
Query: 494 ------PITIQYNPESEPLTLEPELT-IPVQTEPEPRMSDHSVPRASERLVSRTTDTDSS 546
P T P + P+T T +P T P P + + P T T S
Sbjct: 1490 TTTTPPPTTTPSPPMTTPITPPASTTTLPPTTTPSPPTTTTTTPPP--------TTTPSP 1541
Query: 547 SVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLV 606
TPI+ PT+ P+ +T T PSP P P +
Sbjct: 1542 PTTTPITPPTSTTTLPPTTTPSP---------PPTTTTTPPPTTTPSPPTTTTPSPPTIT 1592
Query: 607 DPDEPILANPLHEADPLAQQ---AQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEP 663
P P P P P+ P + ++ SP P T+ P
Sbjct: 1593 TTTPPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPP 1652
Query: 664 HLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVPYTSLRPTS 711
P T T+ + P+SP T P T++ S
Sbjct: 1653 PTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITTTPSPPTTTMTTPS 1700
Score = 51.2 bits (121), Expect = 1e-05
Identities = 76/342 (22%), Positives = 110/342 (31%), Gaps = 34/342 (9%)
Query: 371 PSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKS---VVDSDADLNS 427
PSPP + P + + TPP T S +++ S S +
Sbjct: 1438 PSPP------ITTTTTPLPTTTPSPPISTTTTPPPTTTPSPPTTTPSPPTTTPSPPTTTT 1491
Query: 428 FDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTG 487
P + T P+T PA+ TT P + PSP P PT
Sbjct: 1492 TTPPPTTTPSPPMTTPIT-----PPASTTTLPPTTT------PSP---PTTTTTTPPPT- 1536
Query: 488 PSDPTSPITIQYNPESEPLTLEPELTI---PVQTEPEPRMSDHSVPRASERLVSRTTDTD 544
+ P+ P T P + TL P T P T P + S P + T T
Sbjct: 1537 -TTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPTITTTT 1595
Query: 545 SSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPER 604
TP S PT + P + S + ST T PSP P
Sbjct: 1596 PPPTTTP-SPPTTTTTTPPPTTTPSPPTTTPITPP-TSTTTLPPTTTPSPPPTTTTTPPP 1653
Query: 605 LVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPH 664
P P P + P+ P P P P +++ +P+P S P
Sbjct: 1654 TTTPSPPTTTTP---SPPITTTTTPPPTTT-PSSPITTTPSPPTTTMTTPSPTTTPSSPI 1709
Query: 665 LGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVPYTS 706
+ P T P + + P+ P T ++ +P T+
Sbjct: 1710 TTTTTPSSTTTPSPPPTTMTTPSPTTTPSPPTTTMTTLPPTT 1751
Score = 44.3 bits (103), Expect = 0.001
Identities = 60/278 (21%), Positives = 82/278 (28%), Gaps = 31/278 (11%)
Query: 442 NPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTS------PI 495
+P T P S P TT+ P + S +P P T PS PT+ P
Sbjct: 1478 SPPTTTP-SPPTTTTTTPPPTTTPSPPMTTPITPPASTTTLPPTTTPSPPTTTTTTPPPT 1536
Query: 496 TIQYNPESEPLTLEPELT-IPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISF 554
T P + P+T T +P T P P + + P + TT T S T +
Sbjct: 1537 TTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPTITTTTP 1596
Query: 555 PTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPER-LVDPDEPIL 613
P S P+ + T P P P P + P
Sbjct: 1597 PPTTTPSPPTTTT----------------------TTPPPTTTPSPPTTTPITPPTSTTT 1634
Query: 614 ANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQ 673
P P P P P ++ +P P S P P
Sbjct: 1635 LPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITTTPSPPTT 1694
Query: 674 TCDIGSPQGTSEAHSSNHPTSPETNLSIVPYTSLRPTS 711
T SP T + + T T P T++ S
Sbjct: 1695 TMTTPSPTTTPSSPITTTTTPSSTTTPSPPPTTMTTPS 1732
Score = 36.6 bits (83), Expect = 0.31
Identities = 36/136 (26%), Positives = 46/136 (33%), Gaps = 11/136 (8%)
Query: 387 PTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTL 446
PT P+ + PP T SS +++ S + S P S + +T T
Sbjct: 1660 PTTTTPSPPITTTTTPPPTTTPSSPITTTPSPPTTTMTTPSPTTTPSSPITTTTTPSSTT 1719
Query: 447 IPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPL 506
P P TT P + PSP P M PT S P + T P P
Sbjct: 1720 TPSPPPTTMTTPSPTT------TPSP---PTTTMTTLPPTTTSSPLT--TTPLPPSITPP 1768
Query: 507 TLEPELTIPVQTEPEP 522
T P T T P
Sbjct: 1769 TFSPFSTTTPTTPCVP 1784
Score = 31.6 bits (70), Expect = 9.9
Identities = 51/237 (21%), Positives = 71/237 (29%), Gaps = 50/237 (21%)
Query: 485 PTGPSDPTS-PITIQ--YNPESEPLTLEPELTIPVQTE----PEPRMSDHSVPRASERLV 537
PTG PT+ PIT P P + T P+ T P P + P
Sbjct: 4121 PTGTQTPTTTPITTTTTVTPTPTPTGTQTPTTTPITTTTTVTPTPTPTGTQTPT------ 4174
Query: 538 SRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCP-SPRR 596
TT +++ TP PT P++ S T P +
Sbjct: 4175 --TTPITTTTTVTPTPTPTGTQTGPPTHTS----------------------TAPIAELT 4210
Query: 597 YPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNP 656
P PE ++PL E+ L P P +P
Sbjct: 4211 TSNPPPESSTPQTSRSTSSPLTESTTLLSTLPPAIEMTSTAPPS------------TPTA 4258
Query: 657 LVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVPYTSLRPTSLS 713
TS H + P T G+P + SS+ PT + + PT LS
Sbjct: 4259 PTTTSGGHTLSPPPSTTTSPPGTPTRGTTTGSSSAPTPSTVQTTTTSAWTPTPTPLS 4315
>SON_MOUSE (Q9QX47) SON protein
Length = 2404
Score = 52.4 bits (124), Expect = 5e-06
Identities = 96/452 (21%), Positives = 161/452 (35%), Gaps = 62/452 (13%)
Query: 275 QDMVNAGCVEDLNTIVSDVFTAN-----SLKKMGLVQKKIAPAHEDTSSKIRQRRRKMVL 329
+ + N V+ + ++S V LK+ + ++ + T ++ +K
Sbjct: 54 RSLPNEEIVQKIEEVLSGVLDTELRYKPDLKEASRKSRCVSVQTDPTDEVPTKKSKKHKK 113
Query: 330 EESSEESDVPLTKKKKADQPKRKHDETSKPDDGN-DGDN----DGGPSPPK--------- 375
++ ++ +KK QP+ + DGN + D+ D PS
Sbjct: 114 HKNKKKKKKKEKEKKYKRQPEESESKLKSHHDGNLESDSFLKFDSEPSAAALEHPVRAFG 173
Query: 376 -KKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSS-SKSVVDSDADLNSFDALPI 433
+ + LV++P V Q +ET T+++ S S SV+ ++ P+
Sbjct: 174 LSEASETALVLEPPVVSMEVQESHVLETLKPATKAAELSVVSTSVISEQSEQ------PM 227
Query: 434 SAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTS 493
ML+ S + + P +T+ S + + L+ P S T
Sbjct: 228 PGMLEPSMTKILDSFTAAPVPMSTAALKSPEPVVTMSVEYQKSVLKSLETMPPETSKTTL 287
Query: 494 ---PITIQYNPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYT 550
PI P SE LT+ E P + PEP S P +S V R +
Sbjct: 288 VELPIAKVVEP-SETLTIVSET--PTEVHPEPSPSTMDFPESSTTDVQRLPEQ------- 337
Query: 551 PISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVD--- 607
P+ P+ +ADSS + ES +E+ K L+E T S PGP +++
Sbjct: 338 PVEAPSEIADSSMTRPQES----LELPKTTAVELQE--STVASALELPGPPATSILELQG 391
Query: 608 -PDEPILANPLHEADPLAQQAQP----------DPVQQEPVQPDPEQSVSNHSSVRSPNP 656
P P+ P A P+ + + P P P P P + S P P
Sbjct: 392 PPVTPVPELPGPSATPVPELSGPLSTPVPELPGPPATVVPELPGPSVTPVPQLSQELPGP 451
Query: 657 LVDTS--EPHLGASEPHVQTCDIGSPQGTSEA 686
+ EP EP V ++ S A
Sbjct: 452 PAPSMGLEPPQEVPEPPVMAQELSGVPAVSAA 483
Score = 34.3 bits (77), Expect = 1.5
Identities = 59/277 (21%), Positives = 105/277 (37%), Gaps = 19/277 (6%)
Query: 393 AAQVVRRIETPPRVTRSSVRSSS-KSVVDSDADLNSFDALPISAMLKRST---NPLTLIP 448
A++ V + P R SSV S+ +S V ++ ++ P++ M+ +T P L
Sbjct: 1194 ASEAVMTVPEPAREPESSVTSAPVESAVVAEHEM--VPERPMTYMVSETTMSVEPAVLTS 1251
Query: 449 ESQPAAQTTSQPNSLRSSFFQPSPNEAPLWN--MLQNQPTGPSDPTSPITIQYNPESEPL 506
E+ ++T+ +S+R S S L + +QP S +T+ P +
Sbjct: 1252 EASVISETSETYDSMRPSGHAISEVTMSLLEPAVTISQPAEDSLELPSMTV---PAPSTM 1308
Query: 507 TLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNN 566
T + V P + + + E T + +V P+ VA P
Sbjct: 1309 TTTESPVVAVTEIPPVAVPEPPIMAVPELPTMAVVKTPAVAVPEPL-----VAAPEPPTM 1363
Query: 567 SESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQ 626
+ + V + V+ E L P ++P PIL + P+
Sbjct: 1364 ATPELCSLSVSEPPVAVSELPALADPE-HAITAVSGVSSLEPSVPILEPAVSVLQPVMIV 1422
Query: 627 AQPDPVQQEPVQP--DPEQSVSNHSSVRSPNPLVDTS 661
++P QEP +P VS H+ + SP V++S
Sbjct: 1423 SEPSVPVQEPTVAVSEPAVIVSEHTQITSPEMAVESS 1459
>UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89
(Uncoordinated protein 89)
Length = 6632
Score = 51.6 bits (122), Expect = 9e-06
Identities = 72/378 (19%), Positives = 139/378 (36%), Gaps = 33/378 (8%)
Query: 283 VEDLNTIVSDVFTANSLKKMGLVQKKIAPAHEDTSSKIRQRRRKMVLEESSEESDVPLTK 342
+E T ++ T S + V+K+ P D K ++ K + +EE P+ K
Sbjct: 1504 IEKTETTMTTEMTHESEESRTSVKKEKTPEKVDEKPKSPTKKDKSPEKSITEEIKSPVKK 1563
Query: 343 KKKADQPKRKHDETSKPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTRVE--PAAQVVRRI 400
+K ++ + K +K + + SP KK + +V+ PT+ E P VV +
Sbjct: 1564 EKSPEKVEEKPASPTKKEKSPEKP----ASPTKKSENEVK---SPTKKEKSPEKSVVEEL 1616
Query: 401 ETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQP 460
++P + KS + + + K+ +P + + + P
Sbjct: 1617 KSPKEKSPEKADDKPKSPTKKEKSPEKSATEDVKSPTKKEKSP-------EKVEEKPTSP 1669
Query: 461 NSLRSSFFQPSPNEAPLWNMLQNQP-TGPSDPTSPITIQYNPESEPLTLEPELTIPVQTE 519
SS + + +E + P T P SP + +PE ++ E+ P +
Sbjct: 1670 TKKESSPTKKTDDEVKSPTKKEKSPQTVEEKPASPTKKEKSPEK---SVVEEVKSPKEKS 1726
Query: 520 PEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVAD---SSPSNNSESIRKFME- 575
PE P E+ ++ + S P A+ SP+ S K +
Sbjct: 1727 PEKAEEKPKSPTKKEKSPEKSAAEEVKSPTKKEKSPEKSAEEKPKSPTKKESSPVKMADD 1786
Query: 576 -----VRKEKVSTLEEYYLTCPSPRRYPGPR--PERLVDP--DEPILANPLHEADPLAQQ 626
+KEK E P+ + + E L P E ++P + +++
Sbjct: 1787 EVKSPTKKEKSPEKVEEKPASPTKKEKTPEKSAAEELKSPTKKEKSPSSPTKKTGDESKE 1846
Query: 627 AQPDPVQQEPVQPDPEQS 644
P+ +++P P P++S
Sbjct: 1847 KSPEKPEEKPKSPTPKKS 1864
Score = 48.9 bits (115), Expect = 6e-05
Identities = 87/406 (21%), Positives = 137/406 (33%), Gaps = 45/406 (11%)
Query: 296 ANSLKKMGLVQKKIAPAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHDE 355
A+ KK G K +P + +S ++ + ++EE P K+K P +K
Sbjct: 1394 ASPTKKTGEEVK--SPKEKSPASPTKKEK-----SPAAEEVKSPTKKEKSPSSPTKKEKS 1446
Query: 356 TSKPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSS 415
S P GD SPPK K+ + KP V+ + + + V SS +
Sbjct: 1447 PSSPTK-KTGDEVKEKSPPKSPTKKEKSPEKPEDVKSPVKKEKSPDATNIVEVSSETTIE 1505
Query: 416 KSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEA 475
K+ ++ +++ K T P + S S+ P E
Sbjct: 1506 KTETTMTTEMTHESEESRTSVKKEKTPEKVDEKPKSPTKKDKSPEKSITEEIKSPVKKE- 1564
Query: 476 PLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASER 535
+ P+ PT P S E E+ P + E P S E
Sbjct: 1565 ---KSPEKVEEKPASPTKKEKSPEKPASPTKKSENEVKSPTKKEKSPEKS------VVEE 1615
Query: 536 LVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPR 595
L S + + P S PT S + +E ++ +KEK E T P+ +
Sbjct: 1616 LKSPKEKSPEKADDKPKS-PTKKEKSPEKSATEDVKS--PTKKEKSPEKVEEKPTSPTKK 1672
Query: 596 RYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQP-----DPEQSVSNHSS 650
P + D + P ++ P V+++P P PE+SV
Sbjct: 1673 E---SSPTKKTDDE---------VKSPTKKEKSPQTVEEKPASPTKKEKSPEKSVV--EE 1718
Query: 651 VRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPE 696
V+SP P +P T SP+ S A PT E
Sbjct: 1719 VKSPK----EKSPEKAEEKPKSPTKKEKSPE-KSAAEEVKSPTKKE 1759
Score = 42.4 bits (98), Expect = 0.006
Identities = 117/592 (19%), Positives = 209/592 (34%), Gaps = 85/592 (14%)
Query: 298 SLKKMGLVQKKIAPAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHDETS 357
S+ K+ +V K + D +RR EE +E KKKK+ P +K
Sbjct: 1330 SVSKIEVVSK--TDSQTDVREGTPKRRVSFAEEELPKEVIDSDRKKKKSPSPDKKEKSPE 1387
Query: 358 KPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKS 417
K + + SP KK ++V+ P PA +P + +S KS
Sbjct: 1388 KTE-------EKPASPTKKTGEEVK---SPKEKSPA--------SPTKKEKSPAAEEVKS 1429
Query: 418 VVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPL 477
+ K ++P E P++ T + ++ + SP ++P
Sbjct: 1430 PTKKE---------------KSPSSPTK--KEKSPSSPTKKTGDEVK----EKSPPKSP- 1467
Query: 478 WNMLQNQPTGPSDPTSPITIQYNPESEPLT-LEPELTIPVQTEPEPRMSDHSVPRASERL 536
+ P P D SP+ + +P++ + + E TI + E M+ + E
Sbjct: 1468 -TKKEKSPEKPEDVKSPVKKEKSPDATNIVEVSSETTI---EKTETTMTTEMTHESEESR 1523
Query: 537 VSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRR 596
S + V PT S + +E I+ V+KEK E P+ +
Sbjct: 1524 TSVKKEKTPEKVDEKPKSPTKKDKSPEKSITEEIKS--PVKKEKSPEKVEEKPASPTKKE 1581
Query: 597 YPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNP 656
PE+ P P + P ++ P+ E ++ P++ + + +P
Sbjct: 1582 ---KSPEK---PASPTKKSENEVKSPTKKEKSPEKSVVEELK-SPKEKSPEKADDKPKSP 1634
Query: 657 LVDTSEPHLGASEPHVQTCDIGSP--QGTSEAHSSNHPTSPETNLSIVPYTSLRPTSLSE 714
P A+E D+ SP + S PTSP S PT ++
Sbjct: 1635 TKKEKSPEKSATE------DVKSPTKKEKSPEKVEEKPTSPTKKES-------SPTKKTD 1681
Query: 715 CINVFNREASLMLRNVQGQTDLSENAESVAEKWNSLSTWLVAQV--PVMMQHLTAERDQR 772
E + + + E S +K S +V +V P AE +
Sbjct: 1682 ------DEVKSPTKKEKSPQTVEEKPASPTKKEKSPEKSVVEEVKSPKEKSPEKAEEKPK 1735
Query: 773 IKAAKQRFARRVALHEQQ---QRHKLLEAAEEARRKQEQAEEAARQAAAQAE-QARLEAE 828
K++ + A E + ++ K E + E + K +E++ A E ++ + E
Sbjct: 1736 SPTKKEKSPEKSAAEEVKSPTKKEKSPEKSAEEKPKSPTKKESSPVKMADDEVKSPTKKE 1795
Query: 829 RLEAEAEARRLAPV--VFTPAASASTPAHQAAQNVPSNSTPPSSSRLELMEQ 878
+ + E + +P TP SA+ + S S+P + E E+
Sbjct: 1796 KSPEKVEEKPASPTKKEKTPEKSAAEELKSPTKKEKSPSSPTKKTGDESKEK 1847
>NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 kDa
protein) (140 kDa nucleolar phosphoprotein) (Nopp140)
(Nucleolar and coiled-body phosphoprotein 1)
Length = 704
Score = 51.6 bits (122), Expect = 9e-06
Identities = 60/246 (24%), Positives = 96/246 (38%), Gaps = 49/246 (19%)
Query: 314 EDTSSKIRQRRRKMVLEESSEESDVP-----LTKKKK-ADQPKRKHDETSKPDDGNDGDN 367
ED+SS+ + ++K + +++ S VP L+KK A PK+ +T D D
Sbjct: 278 EDSSSEEEEEQKKPMKKKAGPYSSVPPPSVSLSKKSVGAQSPKKAAAQTQPADSSADSSE 337
Query: 368 DGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNS 427
+ S ++KK + VV T +PA + SSS S DSD+ +
Sbjct: 338 ESDSSSEEEKKTPAKTVVSKTPAKPAP------------VKKKAESSSDS-SDSDSSEDE 384
Query: 428 FDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTG 487
A P+SA + P + PAA+ + P QP G
Sbjct: 385 APAKPVSATKSPLSKP--AVTPKPPAAKAVATP----------------------KQPAG 420
Query: 488 PSD-PTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSR-----TT 541
P S + E E + E E T T P+ R++ + P + R ++
Sbjct: 421 SGQKPQSRKADSSSSEEESSSSEEEATKKSVTTPKARVTAKAAPSLPAKQAPRAGGDSSS 480
Query: 542 DTDSSS 547
D++SSS
Sbjct: 481 DSESSS 486
>YDC9_SCHPO (Q10172) Hypothetical protein C25G10.09c in chromosome I
Length = 1794
Score = 50.4 bits (119), Expect = 2e-05
Identities = 89/375 (23%), Positives = 144/375 (37%), Gaps = 78/375 (20%)
Query: 421 SDADLNSFDALPISAMLKRSTNP---LTLIPESQPAAQTT-SQPNSLRSSFFQPSPN-EA 475
S A +NS + P++A + P + + + PA + S P ++ + P P +
Sbjct: 1259 SPAPVNSATSTPVAAPTAQQIQPGKQASAVSSNVPAVSASISTPPAVVPTVQHPQPTKQI 1318
Query: 476 PLWNMLQNQPTGPSDPTSPITIQYNPESE--PLTLEPELTIPVQTEPEPRMSDHSVPRAS 533
P + T S T+PI Q E++ ++LEP + V T P+P++
Sbjct: 1319 PTAAVKDPSTTSTSFNTAPIPQQAPLENQFSKMSLEPPVRPAVPTSPKPQI--------- 1369
Query: 534 ERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKE---------KVSTL 584
DSS+V+ P P ++ PS+N+ + R R++ VS++
Sbjct: 1370 ---------PDSSNVHAPPP-PVQPMNAMPSHNAVNARPSAPERRDSFGSVSSGSNVSSI 1419
Query: 585 EEYYLTCP---SPRRYPG-----------PRPERLVDPDEPILANPLHEAD-PLAQQAQP 629
E+ T P S PG P P V P +P + A P + A P
Sbjct: 1420 EDETSTMPLKASQPTNPGAPSNHAPQVVPPAPMHAVAPVQPKAPGMVTNAPAPSSAPAPP 1479
Query: 630 DPVQQEP--------------VQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEP----H 671
PV Q P V P SV+ ++ S P +S H+ + P H
Sbjct: 1480 APVSQLPPAVPNVPVPSMIPSVAQQPPSSVAPATAPSSTLPPSQSSFAHVPSPAPPAPQH 1539
Query: 672 VQTCDIGS-PQGTSEAHSSNHPTSPETNL-------SIVPYTSLRPT-SLSECINVFNRE 722
+ S P S H S+ P +P+ + +I P T+L + S S + N
Sbjct: 1540 PSAAALSSAPADNSMPHRSS-PYAPQEPVQKPQAINNIAPATNLGTSQSFSPRMGPVNNS 1598
Query: 723 ASLMLRNVQGQTDLS 737
S + N GQ L+
Sbjct: 1599 GSPLAMNAAGQPSLA 1613
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 49.7 bits (117), Expect = 4e-05
Identities = 68/315 (21%), Positives = 114/315 (35%), Gaps = 29/315 (9%)
Query: 407 TRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSS 466
T+SS +SS S + S + S+ + ST + ES ++ TT+ +S
Sbjct: 208 TKSSTTTSSTSESSTTTSSTSESSTTTSSTSESSTTTSST-SESSTSSSTTAPATPTTTS 266
Query: 467 FFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPV--------QT 518
+ P + + +PT P T+P T + S+ T + +P +
Sbjct: 267 CTKEKPTPPTTTSCTKEKPTPPHHDTTPCTKKKTTTSKTCTKKTTTPVPTPSSSTTESSS 326
Query: 519 EPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRK 578
P P S + +S + S TT++ S+ V TP S T + S+P +S + V
Sbjct: 327 APVPTPSSSTTESSSAPVTSSTTESSSAPVPTP-SSSTTESSSAPVTSSTTESSSAPVTS 385
Query: 579 EKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILAN-------PLHEADPLAQQAQPDP 631
ST E S P P P+ ++ P+ + + A
Sbjct: 386 ---STTES------SSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTS 436
Query: 632 VQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPH---LGASEPHVQTCDIGSPQGTSEAHS 688
E S + SS P P T+E + +S + + +P ++ S
Sbjct: 437 STTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESS 496
Query: 689 SNHPTSPETNLSIVP 703
S TS T S P
Sbjct: 497 SAPVTSSTTESSSAP 511
Score = 46.2 bits (108), Expect = 4e-04
Identities = 88/419 (21%), Positives = 147/419 (35%), Gaps = 37/419 (8%)
Query: 317 SSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDGGPSP-PK 375
SS + V ++E S P+ + + +S P + ++ P P P
Sbjct: 531 SSSTTESSSAPVTSSTTESSSAPVPTPSSSTT-----ESSSTPVTSSTTESSSAPVPTPS 585
Query: 376 KKKKQVRLVVKPTRVEPAAQVVRRIETP-PRVTRSSVRSSSKSVVDSDADLNSFDA-LPI 433
+ PT P++ P P + S+ SSS V S + +S P
Sbjct: 586 SSTTESSSAPVPT---PSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPS 642
Query: 434 SAMLKRSTNPL----------TLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQN 483
S+ + S+ P+ + P P++ TT ++ +S S + AP+ +
Sbjct: 643 SSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTES-SSAPVTSSTTE 701
Query: 484 QPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDT 543
+ P S T + + S P+ T + P P S + +S + S TT++
Sbjct: 702 SSSAPVPTPSSSTTESS--SAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTES 759
Query: 544 DSSSVYTPISFPTNVADSSPSNNSESIRKFME--VRKEKVSTLEEYYLTCPSPRRYPGPR 601
S+ V TP S T + + S S + V ST E S P P
Sbjct: 760 SSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTES------SVAPVPTPS 813
Query: 602 PERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTS 661
+ P + P + + P P S S S +P P +S
Sbjct: 814 SSSNITSSAP-SSTPFSSSTESSSVPVPTPSSSTTESSSAPVSSSTTESSVAPVPTPSSS 872
Query: 662 E---PHLGASEPHVQTCDIGSPQGTSEAHSSNHPTS-PETNLSIVPYTSLRPTSLSECI 716
+S P T + S T SS +P S ET++S T++ PT + +
Sbjct: 873 SNITSSAPSSIPFSSTTESFSTGTTVTPSSSKYPGSQTETSVSSTTETTIVPTKTTTSV 931
Score = 45.1 bits (105), Expect = 9e-04
Identities = 85/401 (21%), Positives = 138/401 (34%), Gaps = 50/401 (12%)
Query: 317 SSKIRQRRRKMVLEESSEESDVPLT----KKKKADQPKRKHDETSKPDDGNDGDNDGGP- 371
SS + V ++E S P+T + A + +S P + ++ P
Sbjct: 399 SSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPV 458
Query: 372 -SPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDA 430
+P + V + E ++ V P + S+ SSS V S + +S
Sbjct: 459 PTPSSSTTESSSAPVTSSTTESSSAPV------PTPSSSTTESSSAPVTSSTTESSSAPV 512
Query: 431 -LPISAMLKRSTNPL------TLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQN 483
P S+ + S+ P T S P +T++ +S S E+ + +
Sbjct: 513 PTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSTPVTSS 572
Query: 484 QPTGPSDPT-SPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTD 542
S P +P + S P+ T + P P S + +S + S TT+
Sbjct: 573 TTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTE 632
Query: 543 TDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRP 602
+ S+ V TP S T + S+P S ST E S P P
Sbjct: 633 SSSAPVPTP-SSSTTESSSAPVPTPSS------------STTES------SSAPVPTPSS 673
Query: 603 ERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSE 662
P+ ++ E+ + PV P P S + SS P P T+E
Sbjct: 674 STTESSSAPVTSSTT-ESSSAPVTSSTTESSSAPV-PTPSSSTTESSSAPVPTPSSSTTE 731
Query: 663 PHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVP 703
+S P + +P ++ SS TS T S P
Sbjct: 732 ---SSSAP------VPTPSSSTTESSSAPVTSSTTESSSAP 763
>SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer
layer protein B) (S-layer protein 1)
Length = 1664
Score = 49.3 bits (116), Expect = 5e-05
Identities = 84/387 (21%), Positives = 131/387 (33%), Gaps = 45/387 (11%)
Query: 333 SEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDGGPSPPKKKKKQVRLVVKPTRVEP 392
SE + P+ +D+P DE + D+ D P+P + PT E
Sbjct: 1091 SETPEEPIPTDTPSDEPTPS-DEPTPSDEPTPSDE---PTPSDE----------PTPSET 1136
Query: 393 AAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQP 452
+ + T S + S SD S + P S P IP P
Sbjct: 1137 PEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTP-------SETPEEPIPTDTP 1189
Query: 453 AAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTL-EPE 511
+ + T S +P+P++ P + ++PT P PI P EP EP
Sbjct: 1190 SDEPTPSDEPTPSD--EPTPSDEPTPS---DEPTPSETPEEPIPTD-TPSDEPTPSDEPT 1243
Query: 512 LTIPVQTEPEPRMSDHSVPRAS-ERLVSRTTDTDS---SSVYTPISFPTNVADSSPSNNS 567
+ EP SD P + E + T +D S TP PT + +PS+
Sbjct: 1244 PSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 1303
Query: 568 ESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQA 627
+ + S E + +P P P E DEP ++ +D
Sbjct: 1304 TPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEP-TPSDEPTPSDEPTPSDEPTPSD 1362
Query: 628 QPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAH 687
+P P + P P ++ + +P P T+ P + G G
Sbjct: 1363 EPTPSDE----PTPSETPEEPTPTTTPTPTPSTT--------PTSGSGGSGGSGGGGGGG 1410
Query: 688 SSNHPTSPETNLSIVPYTSLRPTSLSE 714
PTSP + P ++ PT + E
Sbjct: 1411 GGTVPTSPTPTPTSKPTSTPAPTEIEE 1437
Score = 48.9 bits (115), Expect = 6e-05
Identities = 93/424 (21%), Positives = 142/424 (32%), Gaps = 47/424 (11%)
Query: 311 PAHEDTSSKIRQRRRKMVLEESSEE---SDVPLTKKKKADQPKRKHDETSKPDDGNDGDN 367
P+ E T S + E+ EE +D P + +D+P DE + D+ D
Sbjct: 919 PSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPS-DEPTPSDEPTPSDE 977
Query: 368 DGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPP--RVTRSSVRSSSKSVVDSDADL 425
P+P + T EP E P T S + S SD
Sbjct: 978 ---PTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPT 1034
Query: 426 NSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQ----------PNSLRSSFFQPSPNEA 475
S + P T P IP P+ + T P+ + +P+P+E
Sbjct: 1035 PSDEPTPSDEPTPSET-PEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSET 1093
Query: 476 PLWNMLQNQPTG---------PSDPTSPITIQYNPESEPLTLE-PELTIPVQT-EPEPRM 524
P + + P+ PSD +P + + P EP E PE IP T EP
Sbjct: 1094 PEEPIPTDTPSDEPTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTDTPSDEPTP 1152
Query: 525 SDHSV----PRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKEK 580
SD P S+ T S + PI PT+ P+ + E ++
Sbjct: 1153 SDEPTPSDEPTPSDEPTPSDEPTPSETPEEPI--PTDTPSDEPTPSDEPTPSDEPTPSDE 1210
Query: 581 VSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPD 640
+ +E P+P P DEP ++ +D +P P +
Sbjct: 1211 PTPSDE-----PTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSET 1265
Query: 641 PEQSVSNHSSVRSPNPLVDTSEP-HLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNL 699
PE+ + + P P + EP P + P + E S+ PT ET
Sbjct: 1266 PEEPIPTDTPSDEPTP---SDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPE 1322
Query: 700 SIVP 703
+P
Sbjct: 1323 EPIP 1326
Score = 48.1 bits (113), Expect = 1e-04
Identities = 57/252 (22%), Positives = 90/252 (35%), Gaps = 18/252 (7%)
Query: 459 QPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLE-PELTIPVQ 517
QP ++++ +P P + P + PSD +P + + P EP E PE IP
Sbjct: 761 QPAPIKAASDEPIPTDTPSDEPTPSDEPTPSDEPTP-SDEPTPSDEPTPSETPEEPIPTD 819
Query: 518 T-EPEPRMSDHSV----PRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRK 572
T EP SD P S+ T S + PI PT+ P+ + E
Sbjct: 820 TPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPI--PTDTPSDEPTPSDEPTPS 877
Query: 573 FMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPV 632
++ + +E P+P P DEP ++ +D +P P
Sbjct: 878 DEPTPSDEPTPSDE-----PTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPS 932
Query: 633 QQEPVQPDPEQSVSNHSSVRSPNPLVDTSEP-HLGASEPHVQTCDIGSPQGTSEAHSSNH 691
+ PE+ + + P P + EP P + P + E S+
Sbjct: 933 DEPTPSETPEEPIPTDTPSDEPTP---SDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDE 989
Query: 692 PTSPETNLSIVP 703
PT ET +P
Sbjct: 990 PTPSETPEEPIP 1001
Score = 46.6 bits (109), Expect = 3e-04
Identities = 68/279 (24%), Positives = 96/279 (34%), Gaps = 32/279 (11%)
Query: 448 PESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTG---------PSDPTSPITIQ 498
P +P P+ + +P+P+E P + + P+ PSD +P + +
Sbjct: 784 PSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTP-SDE 842
Query: 499 YNPESEPLTLE-PELTIPVQT-EPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPT 556
P EP E PE IP T EP SD P S TP PT
Sbjct: 843 PTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTP---------------SDEPTPSDEPT 887
Query: 557 NVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGP--RPERLVDPDEPILA 614
+ +PS E E + E P+P P P P P+EPI
Sbjct: 888 PSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPT 947
Query: 615 N-PLHEADPLAQQAQPD-PVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHV 672
+ P E P + D P + P E + S+ + +T E + P
Sbjct: 948 DTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPSD 1007
Query: 673 QTCDIGSPQGTSEAHSSNHPT-SPETNLSIVPYTSLRPT 710
+ P + E S+ PT S E S P S PT
Sbjct: 1008 EPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPT 1046
>FIS2_ARATH (Q9ZNT9) Polycomb group protein
FERTILIZATION-INDEPENDENT SEED 2
Length = 692
Score = 49.3 bits (116), Expect = 5e-05
Identities = 89/414 (21%), Positives = 156/414 (37%), Gaps = 68/414 (16%)
Query: 310 APAHEDTSSKIRQ--RRRKMVLEESSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDN 367
+P +S KI ++ + ESSE VP P R H K + + D+
Sbjct: 130 SPPRAHSSEKISDILTTTQLAIAESSEPK-VPHVNDGNVSSPPRAHSSAEKNESTHVNDD 188
Query: 368 DGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNS 427
D SPP+ + + T V I +PP+ SS ++ S + D D
Sbjct: 189 DDVSSPPRAHSLEKN---ESTHVNE-----DNISSPPK-AHSSKKNESTHMNDEDVSFP- 238
Query: 428 FDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTG 487
P + K +++ LT +QPA S+P R S + A + + QP
Sbjct: 239 ----PRTRSSKETSDILT---TTQPAIVEPSEPKVRRGS--RRKQLYAKRYKARETQP-A 288
Query: 488 PSDPTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERL-VSRTTDTDSS 546
++ + P + N E+ V + PE HS+ +AS+ L ++ +SS
Sbjct: 289 IAESSEPKVLHVNDEN------------VSSPPEA----HSLEKASDILTTTQPAIAESS 332
Query: 547 SVYTPISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLV 606
P NV+ + +++S K+ ST + P+ + ++
Sbjct: 333 EPKVPHVNDENVSSTPRAHSS----------KKNKSTRKNVDNVPSPPKTRSSKKTSDIL 382
Query: 607 DPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVS---NHSSVRSP--------- 654
+P +A ++P + D V P +++ S N ++ SP
Sbjct: 383 TTTQPTIA---ESSEPKVRHVNDDNVSSTPRAHSSKKNKSTRKNDDNIPSPPKTRSSKKT 439
Query: 655 NPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTS---PETNLSIVPYT 705
+ ++ ++P + SEP V + T AHSS S + N S+ P T
Sbjct: 440 SNILTRTQPAIAESEPKVPHVNDDKVSSTPRAHSSKKNKSTHKKDDNASLPPKT 493
>TOLA_ECOLI (P19934) TolA protein
Length = 421
Score = 48.1 bits (113), Expect = 1e-04
Identities = 38/120 (31%), Positives = 61/120 (50%), Gaps = 4/120 (3%)
Query: 761 MMQHLTAERDQRIKAAKQRFARRVA---LHEQQQRHKLLEAAEEARRKQEQAEEAARQAA 817
M + AE + +AA+Q +++ L Q+Q+ + EAA++A KQ+QAEEAA +AA
Sbjct: 83 MKEQQAAEELREKQAAEQERLKQLEKERLAAQEQKKQAEEAAKQAELKQKQAEEAAAKAA 142
Query: 818 AQAEQARLEAERLEAEAEARRLAPVVFTPAASASTPAHQAAQNVPSNSTPPSSSRLELME 877
A A +A+ EA+ AE A++ A A + + A AQ + + E E
Sbjct: 143 ADA-KAKAEADAKAAEEAAKKAAADAKKKAEAEAAKAAAEAQKKAEAAAAALKKKAEAAE 201
Score = 35.4 bits (80), Expect = 0.69
Identities = 27/140 (19%), Positives = 60/140 (42%), Gaps = 10/140 (7%)
Query: 763 QHLTAERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQAEQ 822
Q +A+R + K++ A +Q + L+ E+ R ++ ++ A +AA QAE
Sbjct: 70 QESSAKRSDEQRKMKEQQAAEELREKQAAEQERLKQLEKERLAAQEQKKQAEEAAKQAEL 129
Query: 823 ARLEAE----------RLEAEAEARRLAPVVFTPAASASTPAHQAAQNVPSNSTPPSSSR 872
+ +AE + +AEA+A+ AA A A A + + + +
Sbjct: 130 KQKQAEEAAAKAAADAKAKAEADAKAAEEAAKKAAADAKKKAEAEAAKAAAEAQKKAEAA 189
Query: 873 LELMEQRLDTHESMLQDMKQ 892
++++ + E+ + ++
Sbjct: 190 AAALKKKAEAAEAAAAEARK 209
Score = 32.0 bits (71), Expect = 7.6
Identities = 25/92 (27%), Positives = 42/92 (45%), Gaps = 3/92 (3%)
Query: 789 QQQRHKLLEAAEEARRKQEQAEEAAR--QAAAQAEQARLEAERLEAEAEARRLAPVVFTP 846
Q Q + E+ + K++QA E R QAA Q +LE ERL A+ E ++ A
Sbjct: 68 QSQESSAKRSDEQRKMKEQQAAEELREKQAAEQERLKQLEKERLAAQ-EQKKQAEEAAKQ 126
Query: 847 AASASTPAHQAAQNVPSNSTPPSSSRLELMEQ 878
A A +AA +++ + + + E+
Sbjct: 127 AELKQKQAEEAAAKAAADAKAKAEADAKAAEE 158
>YDF3_SCHPO (Q10475) Probable eukaryotic initiation factor C17C9.03
Length = 1403
Score = 47.4 bits (111), Expect = 2e-04
Identities = 112/571 (19%), Positives = 198/571 (34%), Gaps = 56/571 (9%)
Query: 296 ANSLKKMGLVQKKIAPAHED--TSSKIRQRRRKMVLEESSEESDVP--------LTKKKK 345
AN+ KK K + A + SSK + V ++SSE+S +K
Sbjct: 57 ANAQKKTESTGKITSEADTEKYNSSKSPVNKEGSVEKKSSEKSSTNNKPWRGDNTSKPSA 116
Query: 346 ADQPKRKHDETSKPDDGND-GDNDGGPSPPKKKKKQVRLVVKPTRVEPA--AQVVRRIET 402
+R + KP+ + G ++ P +K + P P + R
Sbjct: 117 NSSAERTSSQHQKPETSSQIGKDNAAPVENVNEKSTSQETAPPVSTVPIQFGSITRNAAI 176
Query: 403 PPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNS 462
P + + S +KS V S + + +++ + P+ PE+ A +P +
Sbjct: 177 PSK-PKVSGNMQNKSGVSSYSSKSQSVNSSVTSNPPHTEEPVAAKPEASSTATKGPRPTT 235
Query: 463 LRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPEP 522
S+ N AP N+P+ + T P T + + P T
Sbjct: 236 SASNTNTSPANGAPT-----NKPSTDINTTDPATQTTQVSASNSPALSGSSTPSNTSSRS 290
Query: 523 RMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVRKE--K 580
+H S + Y+ N +S + N +F R+
Sbjct: 291 NRQNHGNFSEKRHYDRYGNSHPSYNKYSHYQHGFNYNNSGNNRNESGHPRFRNSRRNYNN 350
Query: 581 VSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQ-------AQPDPVQ 633
Y S + P P+ + + PI P+ P Q P+ VQ
Sbjct: 351 QGAYPTYMSNGRSANQSPRNNPQNVNNGSTPIQI-PVSLQTPYGQVYGQPQYIVDPNMVQ 409
Query: 634 QEPV-QPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHP 692
P+ QP V + V P P++ S V + SP ++
Sbjct: 410 YGPILQPG---YVPQYYPVYHQTPYTQNF-PNMSRSGSQVSDQVVESPNSST-------- 457
Query: 693 TSPETNLSIVPYTSLRPTSLSECINVFNREASLMLRNVQGQTDLSENAESVAEKWNSLST 752
SP + + + ++L V + E + +N N+ +
Sbjct: 458 LSPRNGFAPIVKQQKKSSALKIVNPVTHTEVVVPQKNASSPNPSETNSRAETP------- 510
Query: 753 WLVAQVPVMMQHLTAERDQRIKAAKQRFARRVALHEQQQRHK---LLEAAEEARRKQEQA 809
A P + + ++R IK A Q+ + A E +++ + LEA E A+R+ E
Sbjct: 511 --TAAPPQISEEEASQRKDAIKLAIQQRIQEKAEAEAKRKAEEKARLEAEENAKREAE-- 566
Query: 810 EEAARQAAAQAEQARLEAERLEAEAEARRLA 840
E+A R+A +A++ E + EAE +A+R A
Sbjct: 567 EQAKREAEEKAKREAEEKAKREAEEKAKREA 597
Score = 33.5 bits (75), Expect = 2.6
Identities = 34/119 (28%), Positives = 55/119 (45%), Gaps = 8/119 (6%)
Query: 767 AERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQAEQARLE 826
A+R+ KA ++ A A E +++ K EA E+A+R+ E E+A R+A A++ E
Sbjct: 617 AKREAEEKAKRE--AEEKAKREAEEKAKR-EAEEKAKREAE--EKAKREAEENAKREAEE 671
Query: 827 AERLEAEAEARRLAPVVF---TPAASASTPAHQAAQNVPSNSTPPSSSRLELMEQRLDT 882
+ EAE A+R A T + + + N SS+ L E +DT
Sbjct: 672 KAKREAEENAKREAEEKVKRETEENAKRKAEEEGKREADKNPEIKSSAPLASSEANVDT 730
Score = 33.5 bits (75), Expect = 2.6
Identities = 26/74 (35%), Positives = 44/74 (59%), Gaps = 5/74 (6%)
Query: 767 AERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQAEQARLE 826
A+R+ KA ++ A A E +++ K EA E+A+R+ E E+A R+A +A++ E
Sbjct: 609 AKREAEEKAKRE--AEEKAKREAEEKAKR-EAEEKAKREAE--EKAKREAEEKAKREAEE 663
Query: 827 AERLEAEAEARRLA 840
+ EAE +A+R A
Sbjct: 664 NAKREAEEKAKREA 677
>FAU_DROME (Q9VGX3) Anoxia up-regulated protein
Length = 619
Score = 47.0 bits (110), Expect = 2e-04
Identities = 45/130 (34%), Positives = 63/130 (47%), Gaps = 13/130 (10%)
Query: 767 AERDQRIKAAK---QRFARRVALHEQQQRHKLLE-----AAEEARRKQEQAEEAARQAAA 818
AE + +KAA+ Q+ A L E+ K+ E AAEEAR +E A + A + AA
Sbjct: 394 AEEAKALKAAEDAAQKAAEEARLAEEAAAQKVAEEAAQKAAEEARLAEEAAAQKAAEEAA 453
Query: 819 Q--AEQARLEA--ERLEAEAEARRLAPVVFTPAASASTPAHQAAQNVPSNSTPPSSSRLE 874
Q AE+A L+A E AE A++ A A + A +AAQ + +RLE
Sbjct: 454 QKAAEEAALKAAEEARLAEEAAQKAAEEAALKAVEEARAAEEAAQKAAEEARVAEEARLE 513
Query: 875 LMEQRLDTHE 884
EQR+ E
Sbjct: 514 -EEQRVREQE 522
Score = 46.2 bits (108), Expect = 4e-04
Identities = 44/169 (26%), Positives = 74/169 (43%), Gaps = 5/169 (2%)
Query: 722 EASLMLRNVQGQTDLSENAESVAEKWNSLSTWLVAQVPVMMQHLTAERDQRIKAAKQ-RF 780
E + + Q E A+ AE+ L+ AQ A + +KAA++ R
Sbjct: 412 EEARLAEEAAAQKVAEEAAQKAAEEAR-LAEEAAAQKAAEEAAQKAAEEAALKAAEEARL 470
Query: 781 ARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQAEQARLEAERLEAEAEARRLA 840
A A ++ L+A EEAR +E A++AA +A AE+ARLE E+ E E RLA
Sbjct: 471 AEEAA--QKAAEEAALKAVEEARAAEEAAQKAAEEARV-AEEARLEEEQRVREQELERLA 527
Query: 841 PVVFTPAASASTPAHQAAQNVPSNSTPPSSSRLELMEQRLDTHESMLQD 889
+ + A + A+ S + + + +T E ++++
Sbjct: 528 EIEKESEGELARQAAELAEIARQESELAAQELQAIQKNENETSEPVVEE 576
Score = 39.7 bits (91), Expect = 0.037
Identities = 33/105 (31%), Positives = 50/105 (47%), Gaps = 13/105 (12%)
Query: 767 AERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQA---------EEAARQAA 817
A+ QR++ + AL E K + A+EA+R++E+A EA +QAA
Sbjct: 306 AKEKQRLRQERLLTVNEEALDEVDLEKKRAQKADEAKRREERALKEERDRLTAEAEKQAA 365
Query: 818 AQAEQARLEAERLEAE----AEARRLAPVVFTPAASASTPAHQAA 858
A+A++A EA ++ AE AEA A A+ A Q A
Sbjct: 366 AKAKKAAEEAAKIAAEEALLAEAAAQKAAEEAKALKAAEDAAQKA 410
Score = 37.7 bits (86), Expect = 0.14
Identities = 34/119 (28%), Positives = 56/119 (46%), Gaps = 6/119 (5%)
Query: 765 LTAERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQ-AEQA 823
LTAE +++ AAK + A A + L EAA + ++ +A +AA AA + AE+A
Sbjct: 356 LTAEAEKQA-AAKAKKAAEEAAKIAAEEALLAEAAAQKAAEEAKALKAAEDAAQKAAEEA 414
Query: 824 RL----EAERLEAEAEARRLAPVVFTPAASASTPAHQAAQNVPSNSTPPSSSRLELMEQ 878
RL A+++ EA + A+A A +AAQ + ++ L E+
Sbjct: 415 RLAEEAAAQKVAEEAAQKAAEEARLAEEAAAQKAAEEAAQKAAEEAALKAAEEARLAEE 473
>SPEN_DROME (Q8SX83) Split ends protein
Length = 5560
Score = 46.6 bits (109), Expect = 3e-04
Identities = 124/619 (20%), Positives = 231/619 (37%), Gaps = 117/619 (18%)
Query: 327 MVLEESSEESDVPLTKKKKADQPKRKHDETSKPDDGNDGDNDGGPSPPKK-KKKQVRLVV 385
+V+ E EE+ + + A +P +E P+ + + + P P + + + V V+
Sbjct: 3328 LVIAEPDEEAALAAKAIETAGEPASILEE---PEMEPEREAEPDPDPEAEIESEPVVEVL 3384
Query: 386 KPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDA------LPISAMLKR 439
P + A Q ++ E + + +S +D+D + N +A L I ++
Sbjct: 3385 DPEELNKAVQSLKH-EDMMDIKADTPQSERDLQIDTDTEENPDEADSSGPSLKIDETVQS 3443
Query: 440 STNPLTLIPESQPAAQTT---------SQPNSLRSSFFQPSP-NEAPLWNMLQNQPTGPS 489
S++P I + P + T SQP S QPS + P + + P G
Sbjct: 3444 SSSPEKSISNNSPTPRETANIDIPNVESQPKLSNESTPQPSVITKLPFLDTPKTVPAGL- 3502
Query: 490 DPTSPITIQYNPESEPLTLEPELTIPVQTE-PEPRMSDHSVPRAS------ERLVSRTTD 542
P SP+ I+ S+ L+ L PVQT P P + + S ++S ++
Sbjct: 3503 -PPSPVKIEPPTISK---LQQPLVQPVQTVLPAPHSTGSGISANSVINLDLSNVISSCSN 3558
Query: 543 TDSSSVYTPISFPTNVADSSPSNNS---------------ESIRKFMEVRKEKVSTLEEY 587
T ++S S + + S N+ ++IR + + ++ E
Sbjct: 3559 TSAASATASASASISFGSPTASQNAMPQASTPKQGPITPQQAIRTQSLIMQPPTISIPEQ 3618
Query: 588 YLTCPSPRRYPGPRPERLVDPDE-------PILANPLHEADPLAQQA----QPDPVQQEP 636
P+ P+ P P +PLH Q+ Q PV +
Sbjct: 3619 TPHFAVPQMVLSPQSHHPQQPGTYMVGIRAPSPHSPLHSPGRGVAQSRLVGQLSPVGRPM 3678
Query: 637 V-QPDPEQSVSN----HSSVRSP-----NPLVDTSEPHLGASEPHVQTCDIGS------- 679
V QP P+Q V H+ + SP +PL + L +S T + S
Sbjct: 3679 VSQPSPQQQVQQTQQQHALITSPQSSNISPLASPTTRVLSSSNSPT-TSKVNSYQPRNQQ 3737
Query: 680 -PQGTSEAHSSNHPTSPETNLSIVPYTSL-------RPTSLSECINVFNREASLML---- 727
PQ S + T+P+ L +P + PT +S+ + V ++A+
Sbjct: 3738 VPQQPSPKSVAEVQTTPQ--LMTIPLQKMTPIQVPHHPTIISKVVTVQPQQATQSQVASS 3795
Query: 728 ---------RNVQGQTDLSENAESVAEKWNS------LSTWLVAQVPVMMQHLTAER--- 769
+NV ++ V K + + ++ Q+ QH+ ++
Sbjct: 3796 PPLGSLPPHKNVHLNAHQNQQQPQVIAKMTAHQHQQHMQQFMHQQMIQRQQHMQQQQLHG 3855
Query: 770 -DQRIKAAKQRFARRVALHEQQQRH-------KLLEAAEEARRKQEQAEEAARQAAAQAE 821
Q+I +A Q + +QQQ+H + L A + +KQ QA++ Q Q +
Sbjct: 3856 QSQQITSAPQHQMHQQHQAQQQQQHHNQQHLNQQLHAQQHPTQKQHQAQQQFNQQIQQHQ 3915
Query: 822 QARLEAERLEAEAEARRLA 840
+ + + +A+ + L+
Sbjct: 3916 SQQQHQVQQQNQAQQQHLS 3934
Score = 39.3 bits (90), Expect = 0.048
Identities = 35/143 (24%), Positives = 61/143 (42%), Gaps = 29/143 (20%)
Query: 768 ERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQAEQAR--- 824
ERD R K + + R + E++QR K L ++ R ++ + +E +R+A ++ R
Sbjct: 2017 ERDNREKELRDKDLREKEMREKEQREKELHREKDQREREHREKEQSRRAMDVEQEGRGGR 2076
Query: 825 ------LEAERLEAEAEARRLAPV--------------VFTPAASASTPAHQAAQNVPSN 864
+ +++ EA L + TP AS STP + N P
Sbjct: 2077 MRELSSYQKSKMDIAGEASSLTAIDCQHNKENAMDTIAQGTPGASPSTP----SDNTPKE 2132
Query: 865 STPPSS--SRLELMEQRLDTHES 885
+ S S + L ++RL + ES
Sbjct: 2133 RSRKLSRNSPVRLHKRRLSSQES 2155
Score = 38.9 bits (89), Expect = 0.062
Identities = 84/474 (17%), Positives = 166/474 (34%), Gaps = 81/474 (17%)
Query: 391 EPAAQVVRRIETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPES 450
+P Q V+ + P T S + S+ SV++ D N + ++ + + I
Sbjct: 3519 QPLVQPVQTVLPAPHSTGSGI--SANSVINLDLS-NVISSCSNTSAASATASASASISFG 3575
Query: 451 QPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEP 510
P A + P + +P +A L QP PT I PE P P
Sbjct: 3576 SPTASQNAMPQASTPKQGPITPQQAIRTQSLIMQP-----PTISI-----PEQTPHFAVP 3625
Query: 511 ELTIPVQTE--------------PEPRMSDHSVPR--ASERLVSRTTDTDSSSVYTPI-- 552
++ + Q+ P P HS R A RLV + + V P
Sbjct: 3626 QMVLSPQSHHPQQPGTYMVGIRAPSPHSPLHSPGRGVAQSRLVGQLSPVGRPMVSQPSPQ 3685
Query: 553 -----SFPTNVADSSPSNN------SESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPR 601
+ + +SP ++ S + R S + Y + P P+
Sbjct: 3686 QQVQQTQQQHALITSPQSSNISPLASPTTRVLSSSNSPTTSKVNSYQPRNQQVPQQPSPK 3745
Query: 602 PERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTS 661
V ++ PL + P+ P + + V P+Q+ + + S
Sbjct: 3746 SVAEVQTTPQLMTIPLQKMTPIQVPHHPTIISKV-VTVQPQQATQSQVA----------S 3794
Query: 662 EPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVPYTSLRPTSLSECIN--VF 719
P LG+ PH H + H + + + + ++ +
Sbjct: 3795 SPPLGSLPPH------------KNVHLNAHQNQQQPQVIAKMTAHQHQQHMQQFMHQQMI 3842
Query: 720 NREASLMLRNVQGQTD---------LSENAESVAEKWNSLSTWLVAQVPVMMQHLTAERD 770
R+ + + + GQ+ + + ++ ++ + L Q+ QH T ++
Sbjct: 3843 QRQQHMQQQQLHGQSQQITSAPQHQMHQQHQAQQQQQHHNQQHLNQQLHAQ-QHPTQKQH 3901
Query: 771 QRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAAAQAEQAR 824
Q A+Q+F +++ H+ QQ+H++ + + ++ Q + ++Q Q QA+
Sbjct: 3902 Q----AQQQFNQQIQQHQSQQQHQVQQQNQAQQQHLSQQQHQSQQQLNQQHQAQ 3951
>SON_HUMAN (P18583) SON protein (SON3) (Negative regulatory
element-binding protein) (NRE-binding protein) (DBP-5)
(Bax antagonist selected in saccharomyces 1) (BASS1)
Length = 2426
Score = 46.6 bits (109), Expect = 3e-04
Identities = 85/371 (22%), Positives = 134/371 (35%), Gaps = 55/371 (14%)
Query: 301 KMGLVQKKIAPAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKAD-QPKRKHDETSKP 359
K G + + D + +I ++ K + +++ K+KK QP+ +T
Sbjct: 84 KEGSRKSRCVSVQTDPTDEIPTKKSKKHKKHKNKKKKKKKEKEKKYKRQPEESESKTKSH 143
Query: 360 DDGN---DGDN----DGGPSP-----------PKKKKKQVRLVVKPTRVEPAAQVVRRIE 401
DDGN + D+ D PS P + + +V++P V +E
Sbjct: 144 DDGNIDLESDSFLKFDSEPSAVALELPTRAFGPSETNESPAVVLEPPVVSMEVSEPHILE 203
Query: 402 TPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQPN 461
T T+++ S + V S+ S +P +M K I +S AA +
Sbjct: 204 TLKPATKTAELSVVSTSVISEQSEQSVAVMPEPSMTK--------ILDSFAAAPVPTTTL 255
Query: 462 SLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTE-- 519
L+SS +P + + M + S P I LEP T+ V +E
Sbjct: 256 VLKSS--EPVVTMSVEYQMKSVLKSVESTSPEPSKIMLVEPPVAKVLEPSETLVVSSETP 313
Query: 520 ----PEPRMS-DHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFM 574
PEP S P +S R + P+ P+ +ADSS + E
Sbjct: 314 TEVYPEPSTSTTMDFPESSAIEALRLPE-------QPVDVPSEIADSSMTRPQEL----- 361
Query: 575 EVRKEKVSTLEEYYLTCPSPRRYPGP----RPERLVDPDEPILANPLHEADPLAQQAQP- 629
K + LE + S PGP PE P P+L P A P+ + P
Sbjct: 362 -PELPKTTALELQESSVASAMELPGPPATSMPELQGPPVTPVLELPGPSATPVPELPGPL 420
Query: 630 -DPVQQEPVQP 639
PV + P P
Sbjct: 421 STPVPELPGPP 431
>P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K)
Length = 1858
Score = 46.6 bits (109), Expect = 3e-04
Identities = 56/232 (24%), Positives = 98/232 (42%), Gaps = 24/232 (10%)
Query: 353 HDETSKPDDGNDGDNDGGPSPPKKKKKQVR-LVVKPTRVEPAAQVVRRIETPPRVTRSSV 411
+DE ++ N+ N GG S K Q++ L+V + P + V
Sbjct: 254 NDEQFNNNNNNNNSNSGGSSRMITSKSQIKPLIVTSNTAATTTTTTTTNTSAPTTPTNRV 313
Query: 412 RSSSKSVVDS----DADLNSFDALP-ISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSS 466
+SS ++ + +++ + + P ISA+ K+ ++ LIP S + +++ SL SS
Sbjct: 314 QSSLDDLLFNLPTIPSNVPTVNGGPKISAVPKKVSSSKLLIPPSSNVSSSSNITLSLSSS 373
Query: 467 FFQPSPNEA-----PLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPE 521
S + + P+ + + T + P T + S+P T +L IP Q + +
Sbjct: 374 SPSSSSSSSTSTVVPIVQLSSSNSTNSPSTSLPTTPRL---SQPTTSYTQL-IPSQQQQQ 429
Query: 522 PRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKF 573
P S+ S S T T +SS + S ++ PSNNS I F
Sbjct: 430 PTESNSS---------SNTNTTTTSSSSSSSSSSLTISSPQPSNNSIRISAF 472
Score = 35.8 bits (81), Expect = 0.53
Identities = 72/366 (19%), Positives = 141/366 (37%), Gaps = 55/366 (15%)
Query: 400 IETPPRVTRSSVRSSSKSVVDSDADLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQ 459
I + P + SS SS+ +++ + N+ ++ S M+ + LI S AA TT+
Sbjct: 239 ISSSPPPSSSSSSSSNDEQFNNNNNNNNSNSGGSSRMITSKSQIKPLIVTSNTAATTTTT 298
Query: 460 PNSLRSSFFQPSPN-EAPLWNMLQNQPTGPSD-PTSPITIQYNPESEPLTLEPELTIPVQ 517
+ S+ P+ ++ L ++L N PT PS+ PT
Sbjct: 299 TTTNTSAPTTPTNRVQSSLDDLLFNLPTIPSNVPT------------------------- 333
Query: 518 TEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSSPSNNSESIRKFMEVR 577
P++S +S +L+ + SSS +S ++ SS S+++ ++ +++
Sbjct: 334 VNGGPKISAVPKKVSSSKLLIPPSSNVSSSSNITLSLSSSSPSSSSSSSTSTVVPIVQLS 393
Query: 578 KEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHEADPL-AQQAQPDPVQQEP 636
+ L P P L+ P L Q Q P +
Sbjct: 394 SSNSTNSPSTSL------------------PTTPRLSQPTTSYTQLIPSQQQQQPTESNS 435
Query: 637 VQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGSPQGTSEAHSSNHPTSPE 696
+ S+ SS S + + + +P S ++ G S+ P+SP
Sbjct: 436 SSNTNTTTTSSSSSSSSSSLTISSPQP----SNNSIRISAFGRSSTQFTISSNGIPSSP- 490
Query: 697 TNLSIVPYTSLRPTSLSECINVFNREASLM---LRNVQGQTDLSENAESVAEKWNSLSTW 753
+S Y ++ S S V N++ S++ + + ++D+S + S+ NS+
Sbjct: 491 GQVSNKVYNNIGNLSNSSGERVKNKQYSMLNISKKTILDESDISSSPRSIGSP-NSIRAS 549
Query: 754 LVAQVP 759
+ +Q+P
Sbjct: 550 ISSQLP 555
>NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130
kDa protein) (140 kDa nucleolar phosphoprotein)
(Nopp140) (Nucleolar and coiled-body phosphoprotein 1)
Length = 699
Score = 46.6 bits (109), Expect = 3e-04
Identities = 100/542 (18%), Positives = 196/542 (35%), Gaps = 60/542 (11%)
Query: 300 KKMGLVQKKIAPAHEDTSSKIRQRRRKMVLEESSEESDVPLTKKKKADQPKRKHDETSKP 359
K++GL K A ++SS R ++ ++ P+ K K K
Sbjct: 110 KRVGLPPGKAAAKASESSSSEESRDD----DDEEDQKKQPVQKGVKPQAKAAKAPPKKAK 165
Query: 360 DDGNDGDNDGGPSPPKKKKKQVRLVVKPTRVEPAAQVVRRIETPPRVTRSSVRSSSKSVV 419
+D D+ PPK +K ++ V V + + PP+ R++ + ++
Sbjct: 166 SSDSDSDSSSEDEPPKNQKPKITPVT----------VKAQTKAPPKPARAAPKIANGKAA 215
Query: 420 DSDADLNSFDALPISAMLKRSTNPLTLIPESQ-----PAAQTTSQPNSLRSSFFQPSPNE 474
S + +S + S K + P +P+ Q P T+ SS S E
Sbjct: 216 SSSSSSSSSSSSDDSEEEKAAATPKKTVPKKQVVAKAPVKAATTPTRKSSSSEDSSSDEE 275
Query: 475 APLWNMLQNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQ--TEPEPRMSDHSVPRA 532
++N+P GP P + P+ T P+ + Q E SD S +
Sbjct: 276 EEQKKPMKNKP-GPYSYAPPPSAP-PPKKSLGTQPPKKAVEKQQPVESSEDSSDESDSSS 333
Query: 533 SERLVSRTTDTDSSSVYTP---ISFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYL 589
E T S + P + +DSS S++SE + ++ + +
Sbjct: 334 EEEKKPPTKAVVSKATTKPPPAKKAAESSSDSSDSDSSEDDEAPSKPAGTTKNSSNKPAV 393
Query: 590 TCPSPRRYPGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHS 649
T SP P P++ V + +L +AD + + + S+ S
Sbjct: 394 TTKSPAVKPAAAPKQPVGGGQKLLT---RKADSSSSEEE-----------------SSSS 433
Query: 650 SVRSPNPLVDTSEPHLGA----SEPHVQTCDIGSPQGTSEAHSSNHPTSPETNLSIVPYT 705
+V T++P A S P Q +PQG+ ++ S + +S E +
Sbjct: 434 EEEKTKKMVATTKPKATAKAALSLPAKQ-----APQGSRDSSSDSDSSSSEEEEEKTSKS 488
Query: 706 SLRPTSLSECINVFNREASLMLRNVQGQTDLSENAESVAEKWNSLSTWLVAQVPVMMQHL 765
+++ + + + ++ S + +S E+ L + Q
Sbjct: 489 AVKKKPQKVAGGAAPSKPASAKKGKAESSNSSSSDDSSEEEEEKLK----GKGSPRPQAP 544
Query: 766 TAERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQA-AAQAEQAR 824
A + A + A+ E++++ + ++ K+ + EAA++A QA++ +
Sbjct: 545 KANGTSALTAQNGKAAKNSEEEEEEKKKAAVVVSKSGSLKKRKQNEAAKEAETPQAKKIK 604
Query: 825 LE 826
L+
Sbjct: 605 LQ 606
>PODX_RAT (Q9WTQ2) Podocalyxin precursor
Length = 485
Score = 46.2 bits (108), Expect = 4e-04
Identities = 40/144 (27%), Positives = 64/144 (43%), Gaps = 25/144 (17%)
Query: 424 DLNSFDALPISAMLKRSTNPLTLIPESQPAAQTTSQPNSLRSSF--FQPSPNEAPLWNML 481
DLN+ LP + + +P QP + + Q +++SS S N LW +
Sbjct: 142 DLNTSTKLPSTPTTNSTASP------HQPVSHSEGQHTTVQSSSASVSSSDNTTLLWILT 195
Query: 482 QNQPTGPSDPTSPITIQYNPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTT 541
++PTG S+ T PI I P +T PV T +P S P +E + +
Sbjct: 196 TSKPTGTSEGTQPIAIS----------TPGITTPVSTPLQPTGS----PGGTESVPTTEE 241
Query: 542 DTDSSSVYTPISFPTNVADSSPSN 565
T S+S +TP+ + S+PS+
Sbjct: 242 FTHSTSSWTPV---VSQGPSTPSS 262
>DAN4_YEAST (P47179) Cell wall protein DAN4 precursor
Length = 1161
Score = 46.2 bits (108), Expect = 4e-04
Identities = 64/306 (20%), Positives = 106/306 (33%), Gaps = 15/306 (4%)
Query: 440 STNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQY 499
ST P T + P TT ++ ++ + + P + PT + PT+ T
Sbjct: 213 STTPTTSTTSTTPTTSTTPTTSTTSTTSQTSTKSTTPTTSSTSTTPTTSTTPTTSTTSTA 272
Query: 500 NPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVA 559
S T TI S S AS V TT T S++ + + T+ A
Sbjct: 273 PTTSTTSTTSTTSTISTAPTTSTTSSTFSTSSASASSVISTTATTSTTFASLTTPATSTA 332
Query: 560 DSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHE 619
+ + +S S +T + Y++ SP + V E +
Sbjct: 333 STDHTTSSVSTTNAF-TTSATTTTTSDTYISSSSPSQVTSSAEPTTV--SEVTSSVEPTR 389
Query: 620 ADPLAQQAQPDPVQQ--EPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCDI 677
+ + A+P V + V+P V++ + + + + EP + Q
Sbjct: 390 SSQVTSSAEPTTVSEFTSSVEPTRSSQVTSSAEPTTVSEFTSSVEPTRSS-----QVTSS 444
Query: 678 GSPQGTSEAHSSNHPTSPETNLSIVPYTSLRPTSLSECINVFNREASLMLRNVQGQTDLS 737
P SE SS PT S S PT++SE + S + + T +S
Sbjct: 445 AEPTTVSEFTSSVEPTRSSQVTS-----SAEPTTVSEFTSSVEPTRSSQVTSSAEPTTVS 499
Query: 738 ENAESV 743
E SV
Sbjct: 500 EFTSSV 505
Score = 37.7 bits (86), Expect = 0.14
Identities = 58/304 (19%), Positives = 99/304 (32%), Gaps = 20/304 (6%)
Query: 440 STNPLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQY 499
ST P T + P TTS + ++ P+ + T P+ T+ T
Sbjct: 150 STTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTT 209
Query: 500 NPESEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVA 559
+ S T T P + + + + S + + TT + S++ T + T+
Sbjct: 210 STTSTTPTTSTTSTTPTTSTTPTTSTTSTTSQTSTKSTTPTTSSTSTTPTTSTTPTTSTT 269
Query: 560 DSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLHE 619
++P+ ++ S +T T S ++
Sbjct: 270 STAPTTSTTSTTSTTSTISTAPTT-----STTSSTFSTSSASASSVISTTATTSTTFASL 324
Query: 620 ADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSVRSPNPLVDTSEPHLGASEPHVQTCDIGS 679
P A D SVS ++ + TS+ ++ +S P Q
Sbjct: 325 TTPATSTASTD---------HTTSSVSTTNAFTTSATTTTTSDTYISSSSPS-QVTSSAE 374
Query: 680 PQGTSEAHSSNHPTSPETNLSIVPYTSLRPTSLSECINVFNREASLMLRNVQGQTDLSEN 739
P SE SS PT S S PT++SE + S + + T +SE
Sbjct: 375 PTTVSEVTSSVEPTRSSQVTS-----SAEPTTVSEFTSSVEPTRSSQVTSSAEPTTVSEF 429
Query: 740 AESV 743
SV
Sbjct: 430 TSSV 433
>CSP_PLABE (P06915) Circumsporozoite protein precursor (CS)
Length = 339
Score = 45.8 bits (107), Expect = 5e-04
Identities = 50/213 (23%), Positives = 74/213 (34%), Gaps = 43/213 (20%)
Query: 443 PLTLIPESQPAAQTTSQPNSLRSSFFQPSPNEAPLWNMLQNQPTGPSDPTSPITIQYNPE 502
P+ +++ + + N L+ P+PN+ P N P P+DP P NP
Sbjct: 69 PMLRRKKNEKKNEKIERNNKLKQPPPPPNPNDPPPPNPNDPPPPNPNDPPPP-----NPN 123
Query: 503 SEPLTLEPELTIPVQTEPEPRMSDHSVPRASERLVSRTTDTDSSSVYTPISFPTNVADSS 562
P P P P P +D + P A++ D + P P N D +
Sbjct: 124 DPP---PPNANDP----PPPNANDPAPPNANDPAPPNANDPAPPNANDPA--PPNANDPA 174
Query: 563 PSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILAN-----PL 617
P N ++ P+P P P DP P N P
Sbjct: 175 PPNAND-----------------------PAPPNANDPPPPNPNDPAPPQGNNNPQPQPR 211
Query: 618 HEADPLAQ-QAQPDPVQQEPVQPDPEQSVSNHS 649
+ P Q Q QP P Q QP P+ +N++
Sbjct: 212 PQPQPQPQPQPQPQPQPQPRPQPQPQPGGNNNN 244
>BNB_DROME (P29746) Bangles and beads protein
Length = 442
Score = 45.8 bits (107), Expect = 5e-04
Identities = 62/291 (21%), Positives = 110/291 (37%), Gaps = 34/291 (11%)
Query: 598 PGPRPERLVDPDEPILANPLHEADPLAQQAQPDPVQQEPVQPDPEQSVSNHSSV------ 651
P P E + PD A L + L +P P++ E +P +++ ++V
Sbjct: 20 PVPDEEVAIQPDSGAKAELLTKT-VLPTAVEPAPLKPEAEKPAETKTIEAKAAVPEQPDV 78
Query: 652 -RSPNPLVDTSEP-------HLGASEPHVQTCDIGSPQGT----SEAHSSNHPTSPETNL 699
+P+P +EP L +S P V+T + T ++ N P E
Sbjct: 79 NANPSPSTPAAEPAKEINSVELKSSAPDVETAPAIPEKKTLPEEAKPAQENAPVEAEKKQ 138
Query: 700 SIVPYTSLRPT--SLSECINVFNREASLMLRNVQGQTDLSENAESVAEKWNSLSTWLVAQ 757
T PT + + + N + Q + + + K ++ S A+
Sbjct: 139 EKTARTEAEPTVEAQPQATKAIEQAPEAPAANAEVQKQVVDEVKPQEPKIDAKS----AE 194
Query: 758 VPVMMQHLTAERDQRIKAAKQRFARRVALHEQQQRHKLLEAAEEARRKQEQAEEAARQAA 817
P + + AE++ + R R+ EQ+ K AEE + E AA +AA
Sbjct: 195 EPAIPAVVAAEKETPVPEQPAR-QERINEIEQKDAKKDAAVAEEPAKAAEATPTAAPEAA 253
Query: 818 AQAEQARLEAERLEAEAEARRLAPVVFTPAASASTPAHQAAQNVPSNSTPP 868
+++ + A + +PAA+A++PA QAAQ + P
Sbjct: 254 TKSDS--------NIQVIAPEKKSIESSPAAAAASPAAQAAQAKSGEAPKP 296
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.128 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 105,537,025
Number of Sequences: 164201
Number of extensions: 4786481
Number of successful extensions: 25286
Number of sequences better than 10.0: 853
Number of HSP's better than 10.0 without gapping: 148
Number of HSP's successfully gapped in prelim test: 735
Number of HSP's that attempted gapping in prelim test: 20673
Number of HSP's gapped (non-prelim): 3315
length of query: 904
length of database: 59,974,054
effective HSP length: 119
effective length of query: 785
effective length of database: 40,434,135
effective search space: 31740795975
effective search space used: 31740795975
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0125.11


