

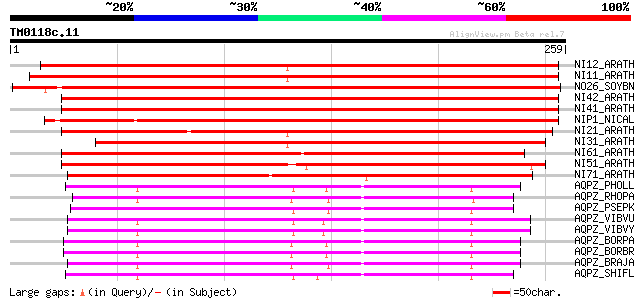
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118c.11
(259 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NI12_ARATH (Q8LFP7) Aquaporin NIP1.2 (NOD26-like intrinsic prote... 363 e-100
NI11_ARATH (Q8VZW1) Aquaporin NIP1.1 (NOD26-like intrinsic prote... 354 1e-97
NO26_SOYBN (P08995) Nodulin-26 (N-26) 349 3e-96
NI42_ARATH (Q8W036) Probable aquaporin NIP4.2 (NOD26-like intrin... 294 2e-79
NI41_ARATH (Q9FIZ9) Putative aquaporin NIP4.1 (NOD26-like intrin... 294 2e-79
NIP1_NICAL (P49173) Probable aquaporin NIP-type (Pollen-specific... 289 5e-78
NI21_ARATH (Q8W037) Aquaporin NIP2.1 (NOD26-like intrinsic prote... 264 1e-70
NI31_ARATH (Q9C6T0) Putative aquaporin NIP3.1 (NOD26-like intrin... 260 3e-69
NI61_ARATH (Q9SAI4) Probable aquaporin NIP6.1 (NOD26-like intrin... 223 3e-58
NI51_ARATH (Q9SV84) Probable aquaporin NIP5.1 (NOD26-like intrin... 216 6e-56
NI71_ARATH (Q8LAI1) Probable aquaporin NIP7.1 (NOD26-like intrin... 168 1e-41
AQPZ_PHOLL (Q7N5C1) Aquaporin Z 148 1e-35
AQPZ_RHOPA (P60925) Aquaporin Z 147 2e-35
AQPZ_PSEPK (Q88F17) Aquaporin Z 147 2e-35
AQPZ_VIBVU (Q8DB17) Aquaporin Z 145 1e-34
AQPZ_VIBVY (Q7MIV9) Aquaporin Z 143 4e-34
AQPZ_BORPA (Q7W917) Aquaporin Z 141 2e-33
AQPZ_BORBR (Q7WKG2) Aquaporin Z 141 2e-33
AQPZ_BRAJA (Q89EG9) Aquaporin Z 140 2e-33
AQPZ_SHIFL (O68874) Aquaporin Z 140 3e-33
>NI12_ARATH (Q8LFP7) Aquaporin NIP1.2 (NOD26-like intrinsic protein
1.2) (Nodulin-26-like major intrinsic protein 2)
(AtNLM2) (NLM2 protein) (NodLikeMip2)
Length = 294
Score = 363 bits (932), Expect = e-100
Identities = 180/249 (72%), Positives = 214/249 (85%), Gaps = 7/249 (2%)
Query: 15 KKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMV 74
KK DS+ VP LQKL+AEVLGTYFLIFAGCA+V VN +DK VTLPGIAIVWGL VMV
Sbjct: 38 KKQDSLLSISVPFLQKLMAEVLGTYFLIFAGCAAVAVNTQHDKAVTLPGIAIVWGLTVMV 97
Query: 75 LVYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIF------ 128
LVYS+GHISGAHFNPAVTIA + RFPLKQVPAY+++QVIGSTLA+ TLRL+F
Sbjct: 98 LVYSLGHISGAHFNPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRLLFGLDQDV 157
Query: 129 -SGKDNHFTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLN 187
SGK + F GTLP+GS+LQ+FV+EFIITF LMFV+SGVATDNRAIGELAGLAVGSTV+LN
Sbjct: 158 CSGKHDVFVGTLPSGSNLQSFVIEFIITFYLMFVISGVATDNRAIGELAGLAVGSTVLLN 217
Query: 188 VLFAGPITGASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNKPVRE 247
V+ AGP++GASMNP RSLGPA+V++ Y+G+WIY+VSPI+GAV+G W Y+ +R T+KP+RE
Sbjct: 218 VIIAGPVSGASMNPGRSLGPAMVYSCYRGLWIYIVSPIVGAVSGAWVYNMVRYTDKPLRE 277
Query: 248 LTKSSSFLK 256
+TKS SFLK
Sbjct: 278 ITKSGSFLK 286
>NI11_ARATH (Q8VZW1) Aquaporin NIP1.1 (NOD26-like intrinsic protein
1.1) (Nodulin-26-like major intrinsic protein 1)
(AtNLM1) (NLM1 protein) (NodLikeMip1)
Length = 296
Score = 354 bits (909), Expect = 1e-97
Identities = 174/254 (68%), Positives = 207/254 (80%), Gaps = 7/254 (2%)
Query: 10 NNEASKKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWG 69
N KK DS+ VP LQKL+AE LGTYFL+F GCASVVVN+ ND VVTLPGIAIVWG
Sbjct: 36 NPRPLKKQDSLLSVSVPFLQKLIAEFLGTYFLVFTGCASVVVNMQNDNVVTLPGIAIVWG 95
Query: 70 LAVMVLVYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIF- 128
L +MVL+YS+GHISGAH NPAVTIA + RFPLKQVPAY+++QVIGSTLA+ TLRL+F
Sbjct: 96 LTIMVLIYSLGHISGAHINPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRLLFG 155
Query: 129 ------SGKDNHFTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGS 182
SGK + F G+ P GSDLQAF +EFI+TF LMF++SGVATDNRAIGELAGLA+GS
Sbjct: 156 LDHDVCSGKHDVFIGSSPVGSDLQAFTMEFIVTFYLMFIISGVATDNRAIGELAGLAIGS 215
Query: 183 TVMLNVLFAGPITGASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITN 242
TV+LNVL A P++ ASMNP RSLGPA+V+ YKGIWIY+V+P LGA+AG W Y+ +R T+
Sbjct: 216 TVLLNVLIAAPVSSASMNPGRSLGPALVYGCYKGIWIYLVAPTLGAIAGAWVYNTVRYTD 275
Query: 243 KPVRELTKSSSFLK 256
KP+RE+TKS SFLK
Sbjct: 276 KPLREITKSGSFLK 289
>NO26_SOYBN (P08995) Nodulin-26 (N-26)
Length = 271
Score = 349 bits (896), Expect = 3e-96
Identities = 167/259 (64%), Positives = 213/259 (81%), Gaps = 5/259 (1%)
Query: 2 ADEVVLNVNNEASK---KCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV 58
+ EVV+NV S+ + DS+ VP LQKLVAE +GTYFLIFAGCAS+VVN N +
Sbjct: 10 SQEVVVNVTKNTSETIQRSDSLVS--VPFLQKLVAEAVGTYFLIFAGCASLVVNENYYNM 67
Query: 59 VTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGST 118
+T PGIAIVWGL + VLVY++GHISG HFNPAVTIA +T+RFPL QVPAY++AQ++GS
Sbjct: 68 ITFPGIAIVWGLVLTVLVYTVGHISGGHFNPAVTIAFASTRRFPLIQVPAYVVAQLLGSI 127
Query: 119 LASGTLRLIFSGKDNHFTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGL 178
LASGTLRL+F G + F+GT+P G++LQAFV EFI+TF LMFV+ GVATDNRA+GE AG+
Sbjct: 128 LASGTLRLLFMGNHDQFSGTVPNGTNLQAFVFEFIMTFFLMFVICGVATDNRAVGEFAGI 187
Query: 179 AVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFI 238
A+GST++LNV+ GP+TGASMNPARSLGPA VH +Y+GIWIY+++P++GA+AG W Y+ +
Sbjct: 188 AIGSTLLLNVIIGGPVTGASMNPARSLGPAFVHGEYEGIWIYLLAPVVGAIAGAWVYNIV 247
Query: 239 RITNKPVRELTKSSSFLKG 257
R T+KP+ E TKS+SFLKG
Sbjct: 248 RYTDKPLSETTKSASFLKG 266
>NI42_ARATH (Q8W036) Probable aquaporin NIP4.2 (NOD26-like intrinsic
protein 4.2) (Nodulin-26-like major intrinsic protein 5)
(AtNLM5) (NLM5 protein) (NodLikeMip5)
Length = 283
Score = 294 bits (752), Expect = 2e-79
Identities = 146/232 (62%), Positives = 179/232 (76%)
Query: 25 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 84
V L QKL+AE++GTYF+IF+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIIFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 85 AHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSD 144
AHFNPAVT+ +RFP QVP YI AQ+ GS LAS TLRL+F+ F GT P S
Sbjct: 99 AHFNPAVTVTFAVFRRFPWYQVPLYIGAQLTGSLLASLTLRLMFNVTPKAFFGTTPTDSS 158
Query: 145 LQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARS 204
QA V E II+FLLMFV+SGVATD+RA GELAG+AVG T++LNV AGPI+GASMNPARS
Sbjct: 159 GQALVAEIIISFLLMFVISGVATDSRATGELAGIAVGMTIILNVFVAGPISGASMNPARS 218
Query: 205 LGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNKPVRELTKSSSFLK 256
LGPAIV +YKGIW+Y+V P +G AG + Y+F+R T+KP+RELTKS+SFL+
Sbjct: 219 LGPAIVMGRYKGIWVYIVGPFVGIFAGGFVYNFMRFTDKPLRELTKSASFLR 270
>NI41_ARATH (Q9FIZ9) Putative aquaporin NIP4.1 (NOD26-like intrinsic
protein 4.1)
Length = 283
Score = 294 bits (752), Expect = 2e-79
Identities = 144/232 (62%), Positives = 178/232 (76%)
Query: 25 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 84
V L QKL+AE++GTYF++F+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIVFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 85 AHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSD 144
AHFNPAVT+ +RFP QVP YI AQ GS LAS TLRL+F F GT PA S
Sbjct: 99 AHFNPAVTVTFAIFRRFPWHQVPLYIGAQFAGSLLASLTLRLMFKVTPEAFFGTTPADSP 158
Query: 145 LQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARS 204
+A V E II+FLLMFV+SGVATDNRA+GELAG+AVG T+M+NV AGPI+GASMNPARS
Sbjct: 159 ARALVAEIIISFLLMFVISGVATDNRAVGELAGIAVGMTIMVNVFVAGPISGASMNPARS 218
Query: 205 LGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNKPVRELTKSSSFLK 256
LGPA+V YK IW+Y+V P+LG ++G + Y+ IR T+KP+RELTKS+SFL+
Sbjct: 219 LGPALVMGVYKHIWVYIVGPVLGVISGGFVYNLIRFTDKPLRELTKSASFLR 270
>NIP1_NICAL (P49173) Probable aquaporin NIP-type (Pollen-specific
membrane integral protein)
Length = 270
Score = 289 bits (739), Expect = 5e-78
Identities = 144/240 (60%), Positives = 184/240 (76%), Gaps = 3/240 (1%)
Query: 17 CDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLV 76
C S+ V +LQKL+AE +GTYF+IFAGC SV VN V T PGI + WGL VMV+V
Sbjct: 33 CSSVS--VVVILQKLIAEAIGTYFVIFAGCGSVAVNKIYGSV-TFPGICVTWGLIVMVMV 89
Query: 77 YSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFT 136
Y++G+ISGAHFNPAVTI RFP KQVP YI+AQ++GS LASGTL L+F +
Sbjct: 90 YTVGYISGAHFNPAVTITFSIFGRFPWKQVPLYIIAQLMGSILASGTLALLFDVTPQAYF 149
Query: 137 GTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITG 196
GT+P GS+ Q+ +E II+FLLMFV+SGVATD+RAIG++AG+AVG T+ LNV AGPI+G
Sbjct: 150 GTVPVGSNGQSLAIEIIISFLLMFVISGVATDDRAIGQVAGIAVGMTITLNVFVAGPISG 209
Query: 197 ASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNKPVRELTKSSSFLK 256
ASMNPARS+GPAIV + Y G+W+Y+V PI+G +AG + Y+ IR T+KP+REL KS+S L+
Sbjct: 210 ASMNPARSIGPAIVKHVYTGLWVYVVGPIIGTLAGAFVYNLIRSTDKPLRELAKSASSLR 269
>NI21_ARATH (Q8W037) Aquaporin NIP2.1 (NOD26-like intrinsic protein
2.1) (Nodulin-26-like major intrinsic protein 4)
(AtNLM4) (NLM4 protein) (NodLikeMip4)
Length = 288
Score = 264 bits (675), Expect = 1e-70
Identities = 137/236 (58%), Positives = 174/236 (73%), Gaps = 8/236 (3%)
Query: 25 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 84
V LQKL+AE++GTY+LIFAGCA++ VN ++ VVTL GIA+VWG+ +MVLVY +GH+S
Sbjct: 44 VHFLQKLLAELVGTYYLIFAGCAAIAVNAQHNHVVTLVGIAVVWGIVIMVLVYCLGHLS- 102
Query: 85 AHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIF-------SGKDNHFTG 137
AHFNPAVT+A +++RFPL QVPAYI QVIGSTLAS TLRL+F S K + F G
Sbjct: 103 AHFNPAVTLALASSQRFPLNQVPAYITVQVIGSTLASATLRLLFDLNNDVCSKKHDVFLG 162
Query: 138 TLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGA 197
+ P+GSDLQAFV+EFIIT LM VV V T R EL GL +G+TV LNV+FAG ++GA
Sbjct: 163 SSPSGSDLQAFVMEFIITGFLMLVVCAVTTTKRTTEELEGLIIGATVTLNVIFAGEVSGA 222
Query: 198 SMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNKPVRELTKSSS 253
SMNPARS+GPA+V YKGIWIY+++P LGAV+G + + E +K+ S
Sbjct: 223 SMNPARSIGPALVWGCYKGIWIYLLAPTLGAVSGALIHKMLPSIQNAEPEFSKTGS 278
>NI31_ARATH (Q9C6T0) Putative aquaporin NIP3.1 (NOD26-like intrinsic
protein 3.1)
Length = 269
Score = 260 bits (664), Expect = 3e-69
Identities = 122/217 (56%), Positives = 166/217 (76%), Gaps = 7/217 (3%)
Query: 41 LIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTIAHVTTKR 100
+IFAGC+++VVN K VTLPGIA+VWGL V V++YSIGH+SGAHFNPAV+IA ++K+
Sbjct: 1 MIFAGCSAIVVNETYGKPVTLPGIALVWGLVVTVMIYSIGHVSGAHFNPAVSIAFASSKK 60
Query: 101 FPLKQVPAYILAQVIGSTLASGTLRLIF-------SGKDNHFTGTLPAGSDLQAFVVEFI 153
FP QVP YI AQ++GSTLA+ LRL+F S K + + GT P+ S+ +FV+EFI
Sbjct: 61 FPFNQVPGYIAAQLLGSTLAAAVLRLVFHLDDDVCSLKGDVYVGTYPSNSNTTSFVMEFI 120
Query: 154 ITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQ 213
TF LMFV+S VATD RA G AG+A+G+T++L++LF+GPI+GASMNPARSLGPA++
Sbjct: 121 ATFNLMFVISAVATDKRATGSFAGIAIGATIVLDILFSGPISGASMNPARSLGPALIWGC 180
Query: 214 YKGIWIYMVSPILGAVAGTWAYSFIRITNKPVRELTK 250
YK +W+Y+VSP++GA++G W Y +R T K E+ +
Sbjct: 181 YKDLWLYIVSPVIGALSGAWTYGLLRSTKKSYSEIIR 217
>NI61_ARATH (Q9SAI4) Probable aquaporin NIP6.1 (NOD26-like intrinsic
protein 6.1)
Length = 305
Score = 223 bits (569), Expect = 3e-58
Identities = 118/216 (54%), Positives = 148/216 (67%), Gaps = 1/216 (0%)
Query: 25 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 84
V L +KL AE +GT LIFAG A+ +VN D TL G A GLAVM+++ S GHISG
Sbjct: 76 VSLYRKLGAEFVGTLILIFAGTATAIVNQKTDGAETLIGCAASAGLAVMIVILSTGHISG 135
Query: 85 AHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSD 144
AH NPAVTIA K FP K VP YI AQV+ S A+ L+ +F + T+P
Sbjct: 136 AHLNPAVTIAFAALKHFPWKHVPVYIGAQVMASVSAAFALKAVFEPTMSGGV-TVPTVGL 194
Query: 145 LQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARS 204
QAF +EFII+F LMFVV+ VATD RA+GELAG+AVG+TVMLN+L AGP T ASMNP R+
Sbjct: 195 SQAFALEFIISFNLMFVVTAVATDTRAVGELAGIAVGATVMLNILIAGPATSASMNPVRT 254
Query: 205 LGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRI 240
LGPAI N Y+ IW+Y+ +PILGA+ G Y+ +++
Sbjct: 255 LGPAIAANNYRAIWVYLTAPILGALIGAGTYTIVKL 290
>NI51_ARATH (Q9SV84) Probable aquaporin NIP5.1 (NOD26-like intrinsic
protein 5.1) (Nodulin-26-like major intrinsic protein 6)
(AtNLM6) (NLM6 protein) (NodLikeMip6)
Length = 304
Score = 216 bits (549), Expect = 6e-56
Identities = 116/234 (49%), Positives = 155/234 (65%), Gaps = 11/234 (4%)
Query: 25 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 84
V L +KL AE +GT+ LIF A +VN D TL G A GLAVM+++ S GHISG
Sbjct: 74 VSLTRKLGAEFVGTFILIFTATAGPIVNQKYDGAETLIGNAACAGLAVMIIILSTGHISG 133
Query: 85 AHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTG--TLPAG 142
AH NP++TIA + FP VPAYI AQV S AS L+ +F +G T+P+
Sbjct: 134 AHLNPSLTIAFAALRHFPWAHVPAYIAAQVSASICASFALKGVFHP---FMSGGVTIPSV 190
Query: 143 SDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPA 202
S QAF +EFIITF+L+FVV+ VATD RA+GELAG+AVG+TVMLN+L AGP TG SMNP
Sbjct: 191 SLGQAFALEFIITFILLFVVTAVATDTRAVGELAGIAVGATVMLNILVAGPSTGGSMNPV 250
Query: 203 RSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITN------KPVRELTK 250
R+LGPA+ Y+ +W+Y+V+P LGA++G Y+ +++ + +PVR +
Sbjct: 251 RTLGPAVASGNYRSLWVYLVAPTLGAISGAAVYTGVKLNDSVTDPPRPVRSFRR 304
>NI71_ARATH (Q8LAI1) Probable aquaporin NIP7.1 (NOD26-like intrinsic
protein 7.1)
Length = 275
Score = 168 bits (425), Expect = 1e-41
Identities = 90/218 (41%), Positives = 135/218 (61%), Gaps = 2/218 (0%)
Query: 28 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 87
L+ ++AE++GT+ L+F+ C + + V L A+ GL+V+V+VYSIGHISGAH
Sbjct: 45 LRIVMAELVGTFILMFSVCGVISSTQLSGGHVGLLEYAVTAGLSVVVVVYSIGHISGAHL 104
Query: 88 NPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSDLQA 147
NP++TIA FP QVP YI AQ +G+T A+ + + G + T PA S + A
Sbjct: 105 NPSITIAFAVFGGFPWSQVPLYITAQTLGATAAT-LVGVSVYGVNADIMATKPALSCVSA 163
Query: 148 FVVEFIITFLLMFVVSGV-ATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARSLG 206
F VE I T +++F+ S + ++ +G L G +G+ + L VL GPI+G SMNPARSLG
Sbjct: 164 FFVELIATSIVVFLASALHCGPHQNLGNLTGFVIGTVISLGVLITGPISGGSMNPARSLG 223
Query: 207 PAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNKP 244
PA+V ++ +WIYM +P++GA+ G Y I + +P
Sbjct: 224 PAVVAWDFEDLWIYMTAPVIGAIIGVLTYRSISLKTRP 261
>AQPZ_PHOLL (Q7N5C1) Aquaporin Z
Length = 231
Score = 148 bits (374), Expect = 1e-35
Identities = 90/227 (39%), Positives = 130/227 (56%), Gaps = 16/227 (7%)
Query: 27 LLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV-VTLPGIAIVWGLAVMVLVYSIGHISGA 85
+L+KL AE+LGT+ L+F GC SVV ++ + G+++ +GL V+ ++Y++GHISG
Sbjct: 1 MLRKLAAELLGTFVLVFGGCGSVVFAAAFPELGIGFVGVSLAFGLTVLTMIYAVGHISGG 60
Query: 86 HFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGK-----------DNH 134
HFNPAVTI RF +V YI++QVIG LA+ L +I SG+ N
Sbjct: 61 HFNPAVTIGLWAGGRFRAVEVIPYIISQVIGGILAAAVLYVIASGQVGFDATTSGFASNG 120
Query: 135 FTGTLPAGSDLQ-AFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGP 193
F P G LQ A V E ++T + + V+ G ATD RA A LA+G ++L L + P
Sbjct: 121 FGEHSPGGFSLQSAIVAEIVLTAIFLIVIIG-ATDRRAPPGFAPLAIGLALVLINLISIP 179
Query: 194 ITGASMNPARSLGPAIVHNQY--KGIWIYMVSPILGAVAGTWAYSFI 238
IT S+NPARS AI N + + +W + V PI+G + G Y +
Sbjct: 180 ITNTSVNPARSTAVAIFQNTWALEQLWFFWVMPIIGGIVGGGIYRLL 226
>AQPZ_RHOPA (P60925) Aquaporin Z
Length = 240
Score = 147 bits (372), Expect = 2e-35
Identities = 85/220 (38%), Positives = 132/220 (59%), Gaps = 15/220 (6%)
Query: 30 KLVAEVLGTYFLIFAGCASVVVNLNNDKV-VTLPGIAIVWGLAVMVLVYSIGHISGAHFN 88
K +AE++GT++L FAGC S V+ +V + L G+++ +GL+V+ + Y+IGHISG H N
Sbjct: 5 KYLAEMIGTFWLTFAGCGSAVIAAGFPQVGIGLVGVSLAFGLSVVTMAYAIGHISGCHLN 64
Query: 89 PAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSG----------KDNHFTGT 138
PAVT+ RFP+KQ+ YI+AQV+G+ A+ L LI SG N +
Sbjct: 65 PAVTLGLAAGGRFPVKQIAPYIIAQVLGAIAAAALLYLIASGAAGFDLAKGFASNGYGAH 124
Query: 139 LPAGSDLQA-FVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGA 197
P +L A FV+E ++T + +FV+ G +T +A A LA+G +++ L + P+T
Sbjct: 125 SPGQYNLVACFVMEVVMTMMFLFVIMG-STHGKAPAGFAPLAIGLALVMIHLVSIPVTNT 183
Query: 198 SMNPARSLGPAIVHNQYK--GIWIYMVSPILGAVAGTWAY 235
S+NPARS GPA+ + +W++ V+P+LG V G Y
Sbjct: 184 SVNPARSTGPALFVGGWAIGQLWLFWVAPLLGGVLGGVIY 223
>AQPZ_PSEPK (Q88F17) Aquaporin Z
Length = 230
Score = 147 bits (372), Expect = 2e-35
Identities = 83/220 (37%), Positives = 131/220 (58%), Gaps = 14/220 (6%)
Query: 29 QKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFN 88
+++ AE++GT++L+ GC S V+ ++ + + G+A +GL V+ + ++IGHISG H N
Sbjct: 5 KRMGAELIGTFWLVLGGCGSAVLAASSPLGIGVLGVAFAFGLTVLTMAFAIGHISGCHLN 64
Query: 89 PAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGK----------DNHFTGT 138
PAV+ V RFP K++ Y++AQVIG+ LA+G + LI SGK N +
Sbjct: 65 PAVSFGLVVGGRFPAKELLPYVIAQVIGAILAAGVIYLIASGKAGFELSAGLASNGYADH 124
Query: 139 LPAGSDLQA-FVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGA 197
P G L A FV E ++T + + V+ G ATD RA A +A+G + L L + P+T
Sbjct: 125 SPGGYTLGAGFVSEVVMTAMFLVVIMG-ATDARAPAGFAPIAIGLALTLIHLISIPVTNT 183
Query: 198 SMNPARSLGPAIVHNQY--KGIWIYMVSPILGAVAGTWAY 235
S+NPARS GPA+ + + +W++ V+P++GA G Y
Sbjct: 184 SVNPARSTGPALFVGGWALQQLWLFWVAPLIGAAIGGALY 223
>AQPZ_VIBVU (Q8DB17) Aquaporin Z
Length = 231
Score = 145 bits (365), Expect = 1e-34
Identities = 89/231 (38%), Positives = 129/231 (55%), Gaps = 16/231 (6%)
Query: 28 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKV-VTLPGIAIVWGLAVMVLVYSIGHISGAH 86
+ K +AE+ GT++L+ GC S V+ V + L G+++ +GL V+ + ++IGHISG H
Sbjct: 1 MNKYLAELFGTFWLVLGGCGSAVLAAAFPDVGIGLLGVSLAFGLTVLTMAFAIGHISGCH 60
Query: 87 FNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGK-----------DNHF 135
NPAVTI RF K++ YILAQVIG +A G L I SG+ N +
Sbjct: 61 LNPAVTIGLWAGGRFEAKEIVPYILAQVIGGVIAGGVLYTIASGQMGFDATSSGFASNGY 120
Query: 136 TGTLPAGSDL-QAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPI 194
P G L A V E ++T + + V+ G ATD RA A +A+G + L L + P+
Sbjct: 121 GEHSPGGYSLTSALVTEVVMTMMFLLVILG-ATDQRAPQGFAPIAIGLCLTLIHLISIPV 179
Query: 195 TGASMNPARSLGPAIVHNQY--KGIWIYMVSPILGAVAGTWAYSFIRITNK 243
T S+NPARS G A+ + +W++ V+PILGA+ G AY I +NK
Sbjct: 180 TNTSVNPARSTGVALYVGDWATAQLWLFWVAPILGALLGAVAYKLISGSNK 230
>AQPZ_VIBVY (Q7MIV9) Aquaporin Z
Length = 231
Score = 143 bits (361), Expect = 4e-34
Identities = 88/231 (38%), Positives = 128/231 (55%), Gaps = 16/231 (6%)
Query: 28 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKV-VTLPGIAIVWGLAVMVLVYSIGHISGAH 86
+ K +AE+ GT++L+ GC S V+ V + L G+++ +GL V+ + ++IGHISG H
Sbjct: 1 MNKYLAELFGTFWLVLGGCGSAVLAAAFPDVGIGLLGVSLAFGLTVLTMAFAIGHISGCH 60
Query: 87 FNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGK-----------DNHF 135
NPAVTI RF K++ YILAQVIG +A G L I SG+ N +
Sbjct: 61 LNPAVTIGLWAGGRFEAKEIVPYILAQVIGGVIAGGVLYTIASGQMGFDATSSGFASNGY 120
Query: 136 TGTLPAGSDL-QAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPI 194
P G L A V E ++ + + V+ G ATD RA A +A+G + L L + P+
Sbjct: 121 GEHSPGGYSLTSALVTEIVMAMMFLLVILG-ATDQRAPQGFAPIAIGLCLTLIHLISIPV 179
Query: 195 TGASMNPARSLGPAIVHNQY--KGIWIYMVSPILGAVAGTWAYSFIRITNK 243
T S+NPARS G A+ + +W++ V+PILGA+ G AY I +NK
Sbjct: 180 TNTSVNPARSTGVALYVGDWATAQLWLFWVAPILGALLGAVAYKLISGSNK 230
>AQPZ_BORPA (Q7W917) Aquaporin Z
Length = 236
Score = 141 bits (355), Expect = 2e-33
Identities = 81/228 (35%), Positives = 128/228 (55%), Gaps = 16/228 (7%)
Query: 26 PLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV-VTLPGIAIVWGLAVMVLVYSIGHISG 84
PL ++ AE GT++L+ GC S V+ +V + G+A+ +GL V+ + Y++GHISG
Sbjct: 3 PLFKRCGAEFFGTFWLVLGGCGSAVLAAGVPQVGIGYAGVALAFGLTVLTMAYAVGHISG 62
Query: 85 AHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSG-----------KDN 133
HFNPAVT+ + RF + VP YI+AQV+G+ +A+ TL I G N
Sbjct: 63 GHFNPAVTVGLAASGRFGWRDVPPYIVAQVVGAIVAAATLASIAQGVAGFDLVASKFAAN 122
Query: 134 HFTGTLPAGSDLQ-AFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAG 192
+ P +Q A + E +++ +FV+ G ATD RA A + +G + L L +
Sbjct: 123 GYGDHSPGKYSMQAALICEIVLSAGFVFVILG-ATDKRAPAGFAPIPIGLALTLIHLISI 181
Query: 193 PITGASMNPARSLGPAIVHNQY--KGIWIYMVSPILGAVAGTWAYSFI 238
P+T S+NPARS GPA+ + + +W++ ++PI GA+ G AY +
Sbjct: 182 PVTNTSVNPARSTGPALFVGGWALEQLWLFWLAPIAGALVGALAYRLV 229
>AQPZ_BORBR (Q7WKG2) Aquaporin Z
Length = 236
Score = 141 bits (355), Expect = 2e-33
Identities = 81/228 (35%), Positives = 128/228 (55%), Gaps = 16/228 (7%)
Query: 26 PLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV-VTLPGIAIVWGLAVMVLVYSIGHISG 84
PL ++ AE GT++L+ GC S V+ +V + G+A+ +GL V+ + Y++GHISG
Sbjct: 3 PLFKRCGAEFFGTFWLVLGGCGSAVLAAGVPQVGIGYAGVALAFGLTVLTMAYAVGHISG 62
Query: 85 AHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSG-----------KDN 133
HFNPAVT+ + RF + VP YI+AQV+G+ +A+ TL I G N
Sbjct: 63 GHFNPAVTVGLAASGRFGWRDVPPYIVAQVVGAIVAAATLASIAQGVAGFDLVASKFAAN 122
Query: 134 HFTGTLPAGSDLQ-AFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAG 192
+ P +Q A + E +++ +FV+ G ATD RA A + +G + L L +
Sbjct: 123 GYGDHSPGKYSMQAALICEIVLSAGFVFVILG-ATDKRAPAGFAPIPIGLALTLIHLISI 181
Query: 193 PITGASMNPARSLGPAIVHNQY--KGIWIYMVSPILGAVAGTWAYSFI 238
P+T S+NPARS GPA+ + + +W++ ++PI GA+ G AY +
Sbjct: 182 PVTNTSVNPARSTGPALFVGGWALEQLWLFWLAPIAGALVGALAYRLV 229
>AQPZ_BRAJA (Q89EG9) Aquaporin Z
Length = 240
Score = 140 bits (354), Expect = 2e-33
Identities = 79/225 (35%), Positives = 129/225 (57%), Gaps = 15/225 (6%)
Query: 28 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKV-VTLPGIAIVWGLAVMVLVYSIGHISGAH 86
++K AE +GT++L FAGC S V+ +V + L G+++ +GL+V+ + Y+IGHISG H
Sbjct: 3 MKKYAAEAIGTFWLTFAGCGSAVIAAGFPQVGIGLVGVSLAFGLSVVTMAYAIGHISGCH 62
Query: 87 FNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSG----------KDNHFT 136
NPAVT+ RFP Q+ Y++AQV G+ +A+ L +I SG N +
Sbjct: 63 LNPAVTVGLAAGGRFPAGQILPYVIAQVCGAIVAAELLYIIASGAPGFDVTKGFASNGYD 122
Query: 137 GTLPAGSDLQA-FVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGPIT 195
P + A F+ E ++T + +F++ G AT RA A LA+G +++ L + P+T
Sbjct: 123 AHSPGQYSMMACFLTEVVMTMMFLFIIMG-ATHGRAPAGFAPLAIGLALVMIHLVSIPVT 181
Query: 196 GASMNPARSLGPAIVHNQY--KGIWIYMVSPILGAVAGTWAYSFI 238
S+NPARS GPA+ + +W++ V+P++G G Y ++
Sbjct: 182 NTSVNPARSTGPALFVGGWAMAQLWLFWVAPLIGGALGGVIYRWL 226
>AQPZ_SHIFL (O68874) Aquaporin Z
Length = 231
Score = 140 bits (353), Expect = 3e-33
Identities = 84/224 (37%), Positives = 125/224 (55%), Gaps = 16/224 (7%)
Query: 27 LLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV-VTLPGIAIVWGLAVMVLVYSIGHISGA 85
+ +KL AE GT++L+F GC S V+ ++ + G+A+ +GL V+ + +++GHISG
Sbjct: 1 MFRKLAAECFGTFWLVFGGCGSAVLAAGFPELGIGFAGVALAFGLTVLTMAFAVGHISGG 60
Query: 86 HFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGK-----------DNH 134
HFNPAVTI RFP K+V Y++AQV+G +A+ L LI SGK N
Sbjct: 61 HFNPAVTIGLWAGGRFPAKEVVGYVIAQVVGGIVAAALLYLIASGKTGFDAAASGFASNG 120
Query: 135 FTGTLPAG-SDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAVGSTVMLNVLFAGP 193
+ P G S L A VVE +++ + V+ G ATD A A +A+G + L L + P
Sbjct: 121 YGEHSPGGYSMLSALVVELVLSAGFLLVIHG-ATDKFAPAGFAPIAIGLALTLIHLISIP 179
Query: 194 ITGASMNPARSLGPAIVHNQY--KGIWIYMVSPILGAVAGTWAY 235
+T S+NPARS AI + + +W + V PI+G + G Y
Sbjct: 180 VTNTSVNPARSTAVAIFQGGWALEQLWFFWVVPIVGGIIGGLIY 223
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.138 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,955,791
Number of Sequences: 164201
Number of extensions: 1123425
Number of successful extensions: 3345
Number of sequences better than 10.0: 167
Number of HSP's better than 10.0 without gapping: 153
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 2773
Number of HSP's gapped (non-prelim): 194
length of query: 259
length of database: 59,974,054
effective HSP length: 108
effective length of query: 151
effective length of database: 42,240,346
effective search space: 6378292246
effective search space used: 6378292246
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0118c.11


