

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118a.3
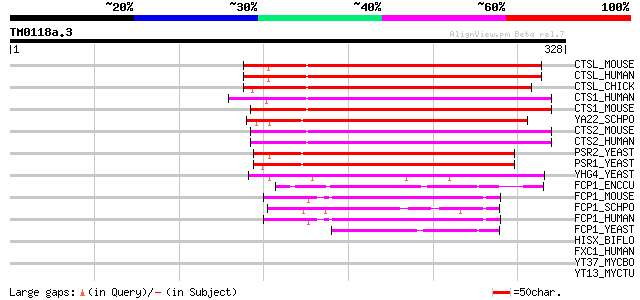
(328 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CTSL_MOUSE (P58465) CTD small phosphatase-like protein (CTDSP-li... 143 5e-34
CTSL_HUMAN (O15194) CTD small phosphatase-like protein (CTDSP-li... 143 5e-34
CTSL_CHICK (Q9PTJ6) CTD small phosphatase-like protein (CTDSP-li... 141 3e-33
CTS1_HUMAN (Q9GZU7) Carboxy-terminal domain RNA polymerase II po... 135 1e-31
CTS1_MOUSE (P58466) Carboxy-terminal domain RNA polymerase II po... 135 1e-31
YA22_SCHPO (Q09695) Hypothetical protein C2F7.02c in chromosome I 133 5e-31
CTS2_MOUSE (Q8BX07) Carboxy-terminal domain RNA polymerase II po... 130 6e-30
CTS2_HUMAN (O14595) Carboxy-terminal domain RNA polymerase II po... 129 1e-29
PSR2_YEAST (Q07949) Probable phosphatase PSR2 (EC 3.1.3.-) (Plas... 122 9e-28
PSR1_YEAST (Q07800) Phosphatase PSR1 (EC 3.1.3.-) (Plasma membra... 122 1e-27
YHG4_YEAST (P38757) Hypothetical 50.6 kDa protein in RPL14B-GPA1... 87 7e-17
FCP1_ENCCU (Q8SV03) RNA polymerase II subunit A C-terminal domai... 62 2e-09
FCP1_MOUSE (Q7TSG2) RNA polymerase II subunit A C-terminal domai... 49 2e-05
FCP1_SCHPO (Q9P376) RNA polymerase II subunit A C-terminal domai... 46 1e-04
FCP1_HUMAN (Q9Y5B0) RNA polymerase II subunit A C-terminal domai... 46 1e-04
FCP1_YEAST (Q03254) RNA polymerase II subunit A C-terminal domai... 44 6e-04
HISX_BIFLO (Q8G4S9) Histidinol dehydrogenase (EC 1.1.1.23) (HDH) 31 3.7
FXC1_HUMAN (Q12948) Forkhead box protein C1 (Forkhead-related pr... 31 3.7
YT37_MYCBO (P65531) Hypothetical protein Mb2937c 31 4.9
YT13_MYCTU (P65530) Hypothetical protein Rv2913c/MT2981 31 4.9
>CTSL_MOUSE (P58465) CTD small phosphatase-like protein (CTDSP-like)
(Small C-terminal domain phosphatase 3) (Nuclear LIM
interactor-interacting factor 1) (NLI-interacting factor
1) (NIF-like protein)
Length = 276
Score = 143 bits (361), Expect = 5e-34
Identities = 81/180 (45%), Positives = 113/180 (62%), Gaps = 5/180 (2%)
Query: 139 PSPPLPLLPPSIS----HRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVK 194
PSPP L P ++ ++ V +DLDETLVHS+ P + DF+V IDG + YV
Sbjct: 88 PSPPAKYLLPEVTVLDYGKKCVVIDLDETLVHSSFKPISNA-DFIVPVEIDGTIHQVYVL 146
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVK 254
KRP VDEFL+ + FE V+FTA+L +YA V D +DR + RL+R+SC G VK
Sbjct: 147 KRPHVDEFLQRMGQLFECVLFTASLAKYADPVADLLDRWGVFRARLFRESCVFHRGNYVK 206
Query: 255 DLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDM 314
DLS GR+L +V+IVD++P S+ PENA+ ++ + DD+ D EL L FF+G+ DD+
Sbjct: 207 DLSRLGRELSKVIIVDNSPASYIFHPENAVPVQSWFDDMTDTELLDLIPFFEGLSREDDV 266
>CTSL_HUMAN (O15194) CTD small phosphatase-like protein (CTDSP-like)
(Small C-terminal domain phosphatase 3) (Small CTD
phosphatase 3) (SCP3) (Nuclear LIM
interactor-interacting factor 1) (NLI-interacting factor
1) (NIF-like protein) (RBSP3) (YA22 protein
Length = 276
Score = 143 bits (361), Expect = 5e-34
Identities = 81/180 (45%), Positives = 113/180 (62%), Gaps = 5/180 (2%)
Query: 139 PSPPLPLLPPSIS----HRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVK 194
PSPP L P ++ ++ V +DLDETLVHS+ P + DF+V IDG + YV
Sbjct: 88 PSPPAKYLLPEVTVLDYGKKCVVIDLDETLVHSSFKPISNA-DFIVPVEIDGTIHQVYVL 146
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVK 254
KRP VDEFL+ + FE V+FTA+L +YA V D +DR + RL+R+SC G VK
Sbjct: 147 KRPHVDEFLQRMGQLFECVLFTASLAKYADPVADLLDRWGVFRARLFRESCVFHRGNYVK 206
Query: 255 DLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDM 314
DLS GR+L +V+IVD++P S+ PENA+ ++ + DD+ D EL L FF+G+ DD+
Sbjct: 207 DLSRLGRELSKVIIVDNSPASYIFHPENAVPVQSWFDDMTDTELLDLIPFFEGLSREDDV 266
>CTSL_CHICK (Q9PTJ6) CTD small phosphatase-like protein (CTDSP-like)
(Small C-terminal domain phosphatase 3) (Nuclear LIM
interactor-interacting factor 1) (NLI-interacting factor
1)
Length = 275
Score = 141 bits (355), Expect = 3e-33
Identities = 81/174 (46%), Positives = 110/174 (62%), Gaps = 5/174 (2%)
Query: 139 PSPP----LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVK 194
PSPP LP L S ++ V +DLDETLVHS+ P + DF+V IDG + YV
Sbjct: 87 PSPPAKYLLPELTASDYGKKCVVIDLDETLVHSSFKPISNA-DFIVPVEIDGTIHQVYVL 145
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVK 254
KRP VDEFL+ + FE V+FTA+L +YA V D +DR + RL+R+SC G VK
Sbjct: 146 KRPHVDEFLQRMGELFECVLFTASLAKYADPVADLLDRWGVFRARLFRESCVFHRGNYVK 205
Query: 255 DLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGV 308
DLS GR+L +V+IVD++P S+ PENA+ ++ + DD+ D EL L FF+G+
Sbjct: 206 DLSRLGRELSKVIIVDNSPASYIFHPENAVPVQSWFDDMTDTELLDLIPFFEGL 259
>CTS1_HUMAN (Q9GZU7) Carboxy-terminal domain RNA polymerase II
polypeptide A small phosphatase 1 (EC 3.1.3.16) (Nuclear
LIM interactor-interacting factor 3) (NLI-interacting
factor 3) (NLI-IF)
Length = 261
Score = 135 bits (341), Expect = 1e-31
Identities = 80/195 (41%), Positives = 115/195 (58%), Gaps = 5/195 (2%)
Query: 130 LVFDGGDNDPSPPLPLLPPSI----SHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVID 185
LV + G P+ L P S + V +DLDETLVHS+ P + DF++ ID
Sbjct: 63 LVEENGAIPKQTPVQYLLPEAKAQDSDKICVVIDLDETLVHSSFKPVNNA-DFIIPVEID 121
Query: 186 GVPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSC 245
GV + YV KRP VDEFL+ + FE V+FTA+L +YA V D +D+ RL+R+SC
Sbjct: 122 GVVHQVYVLKRPHVDEFLQRMGELFECVLFTASLAKYADPVADLLDKWGAFRARLFRESC 181
Query: 246 KQEDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFF 305
G VKDLS GRDL+RV+I+D++P S+ P+NA+ + + D++ D EL L FF
Sbjct: 182 VFHRGNYVKDLSRLGRDLRRVLILDNSPASYVFHPDNAVPVASWFDNMSDTELHDLLPFF 241
Query: 306 DGVDCCDDMRDAVKQ 320
+ + DD+ ++Q
Sbjct: 242 EQLSRVDDVYSVLRQ 256
>CTS1_MOUSE (P58466) Carboxy-terminal domain RNA polymerase II
polypeptide A small phosphatase 1 (EC 3.1.3.16) (Nuclear
LIM interactor-interacting factor 3) (NLI-interacting
factor 3) (Golli-interacting protein) (GIP)
Length = 261
Score = 135 bits (340), Expect = 1e-31
Identities = 76/178 (42%), Positives = 109/178 (60%), Gaps = 1/178 (0%)
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP S + V +DLDETLVHS+ P + DF++ IDGV + YV KRP VDEF
Sbjct: 80 LPEAKAQDSDKICVVIDLDETLVHSSFKPVNNA-DFIIPVEIDGVVHQVYVLKRPHVDEF 138
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
L+ + FE V+FTA+L +YA V D +D+ RL+R+SC G VKDLS GRD
Sbjct: 139 LQRMGELFECVLFTASLAKYADPVADLLDKWGAFRARLFRESCVFHRGNYVKDLSRLGRD 198
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAVKQ 320
L+RV+I+D++P S+ P+NA+ + + D++ D EL L FF+ + DD+ ++Q
Sbjct: 199 LRRVLILDNSPASYVFHPDNAVPVASWFDNMSDTELHDLLPFFEQLSRVDDVYSVLRQ 256
>YA22_SCHPO (Q09695) Hypothetical protein C2F7.02c in chromosome I
Length = 325
Score = 133 bits (335), Expect = 5e-31
Identities = 77/179 (43%), Positives = 110/179 (61%), Gaps = 14/179 (7%)
Query: 141 PPLP----------LLPPSISH---RRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGV 187
PPLP LLPP ++ + LDLDETLVHS+ E DFVV IDG+
Sbjct: 132 PPLPEVKRYGEGNWLLPPIAKEDEGKKCLILDLDETLVHSSFKYI-EPADFVVSIEIDGL 190
Query: 188 PMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQ 247
+ V KRPGVDEFL+ + FE+VVFTA+L +YA VLD +D + ++ HRL+R++C
Sbjct: 191 QHDVRVVKRPGVDEFLKKMGDMFEIVVFTASLAKYADPVLDMLDHSHVIRHRLFREACCN 250
Query: 248 EDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFD 306
+G VKDLS GR+L+ +I+D++P+S+ P +A+ I + +D+ D EL L F +
Sbjct: 251 YEGNFVKDLSQLGRNLEDSIIIDNSPSSYIFHPSHAVPISSWFNDMHDMELIDLIPFLE 309
>CTS2_MOUSE (Q8BX07) Carboxy-terminal domain RNA polymerase II
polypeptide A small phosphatase 2 (EC 3.1.3.16)
Length = 270
Score = 130 bits (326), Expect = 6e-30
Identities = 75/178 (42%), Positives = 107/178 (59%), Gaps = 1/178 (0%)
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP + R V +DLDETLVHS+ P + DF+V I+G + YV KRP VDEF
Sbjct: 90 LPEVTEQDQGRICVVIDLDETLVHSSFKPINNA-DFIVPVEIEGTTHQVYVLKRPYVDEF 148
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
L + FE V+FTA+L +YA V D +DR + RL+R++C G VKDLS GRD
Sbjct: 149 LRRMGELFECVLFTASLAKYADPVTDLLDRCGVFRARLFREACVFHQGCYVKDLSRLGRD 208
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAVKQ 320
L++ VI+D++P S+ PENA+ ++ + DD+ D EL L F+ + DD+ ++ Q
Sbjct: 209 LRKTVILDNSPASYIFHPENAVPVQSWFDDMADTELLNLIPVFEELSGTDDVYTSLGQ 266
>CTS2_HUMAN (O14595) Carboxy-terminal domain RNA polymerase II
polypeptide A small phosphatase 2 (EC 3.1.3.16) (Small
CTD phosphatase 2) (SCP2) (Nuclear LIM
interactor-interacting factor 2) (NLI-interacting factor
2) (Protein OS-4)
Length = 271
Score = 129 bits (323), Expect = 1e-29
Identities = 74/178 (41%), Positives = 107/178 (59%), Gaps = 1/178 (0%)
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP + R V +DLDETLVHS+ P + DF+V I+G + YV KRP VDEF
Sbjct: 91 LPEVTEEDQGRICVVIDLDETLVHSSFKPINNA-DFIVPIEIEGTTHQVYVLKRPYVDEF 149
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
L + FE V+FTA+L +YA V D +DR + RL+R+SC G VKDLS GRD
Sbjct: 150 LRRMGELFECVLFTASLAKYADPVTDLLDRCGVFRARLFRESCVFHQGCYVKDLSRLGRD 209
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAVKQ 320
L++ +I+D++P S+ PENA+ ++ + DD+ D EL L F+ + +D+ ++ Q
Sbjct: 210 LRKTLILDNSPASYIFHPENAVPVQSWFDDMADTELLNLIPIFEELSGAEDVYTSLGQ 267
>PSR2_YEAST (Q07949) Probable phosphatase PSR2 (EC 3.1.3.-) (Plasma
membrane sodium response protein 2)
Length = 397
Score = 122 bits (307), Expect = 9e-28
Identities = 67/157 (42%), Positives = 97/157 (61%), Gaps = 4/157 (2%)
Query: 145 LLPPSISH---RRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDE 201
LLPP + ++ + LDLDETLVHS+ S DFV+ ID YV KRPGVDE
Sbjct: 216 LLPPKLQEFQQKKCLILDLDETLVHSSFKYM-HSADFVLPVEIDDQVHNVYVIKRPGVDE 274
Query: 202 FLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGR 261
FL ++ +EVVVFTA++ YA+ +LD +D N + HRL+R++C +G +K+LS GR
Sbjct: 275 FLNRVSQLYEVVVFTASVSRYANPLLDTLDPNGTIHHRLFREACYNYEGNYIKNLSQIGR 334
Query: 262 DLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDREL 298
L +I+D++P S+ P++A+ I + D D EL
Sbjct: 335 PLSETIILDNSPASYIFHPQHAVPISSWFSDTHDNEL 371
>PSR1_YEAST (Q07800) Phosphatase PSR1 (EC 3.1.3.-) (Plasma membrane
sodium response protein 1)
Length = 427
Score = 122 bits (306), Expect = 1e-27
Identities = 68/157 (43%), Positives = 98/157 (62%), Gaps = 4/157 (2%)
Query: 145 LLPP---SISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDE 201
LLPP S ++ + LDLDETLVHS+ S DFV+ ID YV KRPGV+E
Sbjct: 246 LLPPQDESTKGKKCLILDLDETLVHSSFKYL-RSADFVLSVEIDDQVHNVYVIKRPGVEE 304
Query: 202 FLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGR 261
FLE + FEVVVFTA++ Y +LD +D ++++ HRL+R++C +G +K+LS GR
Sbjct: 305 FLERVGKLFEVVVFTASVSRYGDPLLDILDTDKVIHHRLFREACYNYEGNYIKNLSQIGR 364
Query: 262 DLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDREL 298
L ++I+D++P S+ P++AI I + D D EL
Sbjct: 365 PLSDIIILDNSPASYIFHPQHAIPISSWFSDTHDNEL 401
>YHG4_YEAST (P38757) Hypothetical 50.6 kDa protein in RPL14B-GPA1
intergenic region
Length = 446
Score = 86.7 bits (213), Expect = 7e-17
Identities = 57/196 (29%), Positives = 103/196 (52%), Gaps = 21/196 (10%)
Query: 142 PLPLLPPSISH---RRTVFLDLDETLVHSNTAPPPESFD-----FVVRPVIDGVPMEFYV 193
P L+P S+ + ++ + +DLDETL+HS + S V+ + G+ +++
Sbjct: 237 PKKLIPKSVLNTQKKKKLVIDLDETLIHSASRSTTHSNSSQGHLVEVKFGLSGIRTLYFI 296
Query: 194 KKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRN--RLVSHRLYRDSCKQEDG- 250
KRP D FL ++ +++++FTA+++EYA V+D ++ + S R YR C DG
Sbjct: 297 HKRPYCDLFLTKVSKWYDLIIFTASMKEYADPVIDWLESSFPSSFSKRYYRSDCVLRDGV 356
Query: 251 KLVKDLSL----------AGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWK 300
+KDLS+ + L V+I+D++P S+A +NAI + ++ D D +L
Sbjct: 357 GYIKDLSIVKDSEENGKGSSSSLDDVIIIDNSPVSYAMNVDNAIQVEGWISDPTDTDLLN 416
Query: 301 LRMFFDGVDCCDDMRD 316
L F + + D+R+
Sbjct: 417 LLPFLEAMRYSTDVRN 432
>FCP1_ENCCU (Q8SV03) RNA polymerase II subunit A C-terminal domain
phosphatase (EC 3.1.3.16) (CTD phosphatase FCP1)
Length = 411
Score = 62.0 bits (149), Expect = 2e-09
Identities = 54/161 (33%), Positives = 83/161 (51%), Gaps = 24/161 (14%)
Query: 158 LDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVVFTA 217
LDLD+T++H T S + V+ VID + VK RP +D L ++ +E+ V+T
Sbjct: 65 LDLDQTVLH--TTYGTSSLEGTVKFVIDRC--RYCVKLRPNLDYMLRRISKLYEIHVYTM 120
Query: 218 ALREYASLVLDRVD-RNRLVSHRLY-RDSCKQEDGKLVKDLS-LAGRDLKRVVIVDDNPN 274
R YA +++ +D + R+ RD + G LVK LS L D + +VI+DD P+
Sbjct: 121 GTRAYAERIVEIIDPSGKYFDDRIITRD---ENQGVLVKRLSRLFPHDHRNIVILDDRPD 177
Query: 275 SFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMR 315
+ + EN +LIRPF W +F+ VD D +R
Sbjct: 178 VW-DYCENLVLIRPF---------W----YFNRVDINDPLR 204
>FCP1_MOUSE (Q7TSG2) RNA polymerase II subunit A C-terminal domain
phosphatase (EC 3.1.3.16) (TFIIF-associating CTD
phosphatase)
Length = 960
Score = 48.9 bits (115), Expect = 2e-05
Identities = 44/147 (29%), Positives = 69/147 (46%), Gaps = 12/147 (8%)
Query: 151 SHRRTVFLDLDETLVHSNTAPPPES-----FDFVVRPVIDGVPMEFYVKKRPGVDEFLEA 205
+ + + +DLD+TL+H+ P+ F F + G PM + + RP +FLE
Sbjct: 180 NRKLVLMVDLDQTLIHTTEQHCPQMSNKGIFHFQLGR---GEPM-LHTRLRPHCKDFLEK 235
Query: 206 LAAKFEVVVFTAALREYASLVLDRVD-RNRLVSHR-LYRDSCKQEDGKLVKDLSLAGRDL 263
+A +E+ VFT R YA + +D +L SHR L RD C K +L
Sbjct: 236 IAKLYELHVFTFGSRLYAHTIAGFLDPEKKLFSHRILSRDECIDPFSKTGNLRNLFPCGD 295
Query: 264 KRVVIVDDNPNSFANQPENAILIRPFV 290
V I+DD + + P N I ++ +V
Sbjct: 296 SMVCIIDDREDVWKFAP-NLITVKKYV 321
>FCP1_SCHPO (Q9P376) RNA polymerase II subunit A C-terminal domain
phosphatase (EC 3.1.3.16) (CTD phosphatase fcp1)
Length = 723
Score = 45.8 bits (107), Expect = 1e-04
Identities = 41/161 (25%), Positives = 77/161 (47%), Gaps = 33/161 (20%)
Query: 153 RRTVFLDLDETLVHSNTAPP-------PESFDF-VVRPVID--------GVPMEFYVKKR 196
R ++ +DLD+T++H+ P P + ++ V+R V G +Y+K R
Sbjct: 164 RLSLIVDLDQTIIHATVDPTVGEWMSDPGNVNYDVLRDVRSFNLQEGPSGYTSCYYIKFR 223
Query: 197 PGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDL 256
PG+ +FL+ ++ +E+ ++T + YA V +D + +L++D D
Sbjct: 224 PGLAQFLQKISELYELHIYTMGTKAYAKEVAKIID----PTGKLFQDRVLSRD----DSG 275
Query: 257 SLAGRDLKR--------VVIVDDNPNSFANQPENAILIRPF 289
SLA + L+R VV++DD + + P N I + P+
Sbjct: 276 SLAQKSLRRLFPCDTSMVVVIDDRGDVWDWNP-NLIKVVPY 315
>FCP1_HUMAN (Q9Y5B0) RNA polymerase II subunit A C-terminal domain
phosphatase (EC 3.1.3.16) (TFIIF-associating CTD
phosphatase)
Length = 961
Score = 45.8 bits (107), Expect = 1e-04
Identities = 43/147 (29%), Positives = 68/147 (46%), Gaps = 12/147 (8%)
Query: 151 SHRRTVFLDLDETLVHSNTAPPPES-----FDFVVRPVIDGVPMEFYVKKRPGVDEFLEA 205
+ + + +DLD+TL+H+ + F F + G PM + + RP +FLE
Sbjct: 180 NRKLVLMVDLDQTLIHTTEQHCQQMSNKGIFHFQLGR---GEPM-LHTRLRPHCKDFLEK 235
Query: 206 LAAKFEVVVFTAALREYASLVLDRVD-RNRLVSHR-LYRDSCKQEDGKLVKDLSLAGRDL 263
+A +E+ VFT R YA + +D +L SHR L RD C K +L
Sbjct: 236 IAKLYELHVFTFGSRLYAHTIAGFLDPEKKLFSHRILSRDECIDPFSKTGNLRNLFPCGD 295
Query: 264 KRVVIVDDNPNSFANQPENAILIRPFV 290
V I+DD + + P N I ++ +V
Sbjct: 296 SMVCIIDDREDVWKFAP-NLITVKKYV 321
>FCP1_YEAST (Q03254) RNA polymerase II subunit A C-terminal domain
phosphatase (EC 3.1.3.16) (CTD phosphatase FCP1)
Length = 732
Score = 43.9 bits (102), Expect = 6e-04
Identities = 35/102 (34%), Positives = 52/102 (50%), Gaps = 7/102 (6%)
Query: 191 FYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVD-RNRLVSHRLYRDSCKQED 249
+YVK RPG+ EF +A FE+ ++T A R YA + VD L R+ + E+
Sbjct: 245 YYVKVRPGLKEFFAKVAPLFEMHIYTMATRAYALQIAKIVDPTGELFGDRIL---SRDEN 301
Query: 250 GKL-VKDLS-LAGRDLKRVVIVDDNPNSFANQPENAILIRPF 289
G L K L+ L D VV++DD + + N N I + P+
Sbjct: 302 GSLTTKSLAKLFPTDQSMVVVIDDRGDVW-NWCPNLIKVVPY 342
>HISX_BIFLO (Q8G4S9) Histidinol dehydrogenase (EC 1.1.1.23) (HDH)
Length = 471
Score = 31.2 bits (69), Expect = 3.7
Identities = 17/58 (29%), Positives = 30/58 (51%)
Query: 252 LVKDLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVD 309
+++ + L G++L R ++ P + E L+RP +DDV +R LR F + D
Sbjct: 11 IMRIIDLRGQNLSRAELLAAMPRAAMGTSEATDLVRPILDDVKERGAAALRDFEEKFD 68
>FXC1_HUMAN (Q12948) Forkhead box protein C1 (Forkhead-related
protein FKHL7) (Forkhead-related transcription factor 3)
(FREAC-3)
Length = 553
Score = 31.2 bits (69), Expect = 3.7
Identities = 26/94 (27%), Positives = 35/94 (36%), Gaps = 11/94 (11%)
Query: 55 PTKSIKDIRRNNNRRWRRKTPIKNAVAASSAVLASIHRRITKFFSKLARLSPDRKRASYK 114
P I+DI+ N P+ A A S A++ + SPD +S
Sbjct: 219 PPVRIQDIKTENGTCPSPPQPLSPAAALGSGSAAAVPKIE----------SPDSSSSSLS 268
Query: 115 ILKKTSPQDDSICRTLVFDGGDNDPSPPLPLLPP 148
+ P R L DG D+ P PP P PP
Sbjct: 269 S-GSSPPGSLPSARPLSLDGADSAPPPPAPSAPP 301
>YT37_MYCBO (P65531) Hypothetical protein Mb2937c
Length = 611
Score = 30.8 bits (68), Expect = 4.9
Identities = 35/120 (29%), Positives = 52/120 (43%), Gaps = 21/120 (17%)
Query: 187 VPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCK 246
VP E Y G+D L E TAAL + D++ RN L++ R YR S +
Sbjct: 326 VPFELY---SDGID-----LPVFEEFGAGTAALH-----LRDQLQRNELLADRSYRRSFR 372
Query: 247 QEDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFD 306
+E ++ SL RD VIV+ P+ +++ + F +R L L F D
Sbjct: 373 REFDRIKLGPSLWHRDFHDAVIVE--------CPDKSLIGKSFGAIADERGLHPLDAFLD 424
>YT13_MYCTU (P65530) Hypothetical protein Rv2913c/MT2981
Length = 611
Score = 30.8 bits (68), Expect = 4.9
Identities = 35/120 (29%), Positives = 52/120 (43%), Gaps = 21/120 (17%)
Query: 187 VPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCK 246
VP E Y G+D L E TAAL + D++ RN L++ R YR S +
Sbjct: 326 VPFELY---SDGID-----LPVFEEFGAGTAALH-----LRDQLQRNELLADRSYRRSFR 372
Query: 247 QEDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFD 306
+E ++ SL RD VIV+ P+ +++ + F +R L L F D
Sbjct: 373 REFDRIKLGPSLWHRDFHDAVIVE--------CPDKSLIGKSFGAIADERGLHPLDAFLD 424
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,510,555
Number of Sequences: 164201
Number of extensions: 1678902
Number of successful extensions: 4064
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 4027
Number of HSP's gapped (non-prelim): 28
length of query: 328
length of database: 59,974,054
effective HSP length: 110
effective length of query: 218
effective length of database: 41,911,944
effective search space: 9136803792
effective search space used: 9136803792
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0118a.3


