

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0117b.4
(482 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
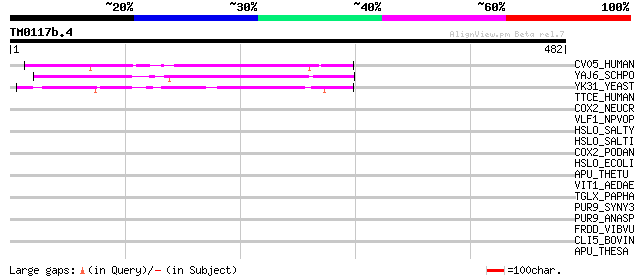
CV05_HUMAN (Q9Y519) Putative MAP kinase activating protein C22orf5 141 4e-33
YAJ6_SCHPO (Q09906) Hypothetical UPF0206 protein C30D11.06c in c... 117 7e-26
YK31_YEAST (P36142) Hypothetical UPF0206 protein YKR051w 115 3e-25
TTCE_HUMAN (Q96N46) Tetratricopeptide repeat protein 14 (TPR rep... 32 2.8
COX2_NEUCR (P00411) Cytochrome c oxidase polypeptide II (EC 1.9.... 32 2.8
VLF1_NPVOP (O10330) Very late expression factor 1 32 3.6
HSLO_SALTY (Q8ZLJ4) 33 kDa chaperonin (Heat shock protein 33 hom... 32 3.6
HSLO_SALTI (Q8Z214) 33 kDa chaperonin (Heat shock protein 33 hom... 32 3.6
COX2_PODAN (P20682) Cytochrome c oxidase polypeptide II (EC 1.9.... 32 3.6
HSLO_ECOLI (P45803) 33 kDa chaperonin (Heat shock protein 33) (H... 32 4.8
APU_THETU (P38536) Amylopullulanase precursor (Alpha-amylase/pul... 32 4.8
VIT1_AEDAE (Q16927) Vitellogenin A1 precursor (VG) (PVG1) [Conta... 31 6.2
TGLX_PAPHA (Q8MIC2) Homeobox protein TGIF2LX (TGFB-induced facto... 31 6.2
PUR9_SYNY3 (P74741) Bifunctional purine biosynthesis protein pur... 31 8.1
PUR9_ANASP (Q8YSJ2) Bifunctional purine biosynthesis protein pur... 31 8.1
FRDD_VIBVU (Q8DCX4) Fumarate reductase subunit D 31 8.1
CLI5_BOVIN (P35526) Chloride intracellular channel protein 5 (Ch... 31 8.1
APU_THESA (P36905) Amylopullulanase precursor (Alpha-amylase/pul... 31 8.1
>CV05_HUMAN (Q9Y519) Putative MAP kinase activating protein C22orf5
Length = 407
Score = 141 bits (355), Expect = 4e-33
Identities = 86/294 (29%), Positives = 150/294 (50%), Gaps = 29/294 (9%)
Query: 14 ATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLV---NP 70
A ++ F+ L ++ + ++ HL Y P EQ++++ ++ +VP Y+ +S++SL+ N
Sbjct: 46 AQAISGFFVWTALLITCHQIYMHLRCYSCPNEQRYIVRILFIVPIYAFDSWLSLLFFTND 105
Query: 71 SISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDY 130
V G +RDCYE+ +Y F LGGE + E +G+ + +
Sbjct: 106 QYYVYFGTVRDCYEALVIYNFLSLCYEYLGGESSIMS--EIRGKPIESSCMY-------- 155
Query: 131 KGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFK 190
GT L + F + K +Q+ ++K ++ V+L+AFG Y +G+F
Sbjct: 156 -GTC--------CLWGKTYSIGFLRFCKQATLQFCVVKPLMAVSTVVLQAFGKYRDGDFD 206
Query: 191 LGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAI 250
+ GY Y+ ++ N S S ALY L FY T++ L+ P+ KF KS++FL++WQG+ +
Sbjct: 207 VTSGYLYVTIIYNISVSLALYALFLFYFATRELLSPYSPVLKFFMVKSVIFLSFWQGMLL 266
Query: 251 ALLYTFGLF------KSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPY 298
A+L G + + +G + + QDFIIC+EM A++ + F K Y
Sbjct: 267 AILEKCGAIPKIHSARVSVGEGTVA-AGYQDFIICVEMFFAALALRHAFTYKVY 319
>YAJ6_SCHPO (Q09906) Hypothetical UPF0206 protein C30D11.06c in
chromosome I
Length = 426
Score = 117 bits (293), Expect = 7e-26
Identities = 82/283 (28%), Positives = 135/283 (46%), Gaps = 28/283 (9%)
Query: 21 FLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSISVDCGILR 80
F+ + L LS + HL YK P Q+ ++ +++M+ YS SF+S+ N I R
Sbjct: 12 FVAIALVLSCISIITHLKNYKKPVLQRSVVRILMMIVIYSSVSFLSVYNEKIGSIFEPFR 71
Query: 81 DCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDYKGTVNHP--F 138
+ YE+FA+YCF L+ LGGE + + LH H + P +
Sbjct: 72 EIYEAFALYCFFCLLIDYLGGERAAV--------------ISLHGH-------LPRPRLW 110
Query: 139 PLKYFLKPWILTRRF-YQIVKFGIVQYMIIKSFTSIMAVILEAFGVY-CEGEFKLGCGYP 196
PL Y L+ + + +K GI+QY +K F I ++ + GVY E +
Sbjct: 111 PLNYLQDDIDLSDPYTFLSIKRGILQYTWLKPFLVIAVLLTKVTGVYDREDQPVYASADL 170
Query: 197 YIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLYTF 256
+I +V N S + +LY L F+ +ELA +P KFL+ K+I+F ++WQ +++
Sbjct: 171 WIGLVYNISITLSLYSLTTFWVCLHEELAPFRPFPKFLSVKAIIFASYWQQTVLSITNWL 230
Query: 257 GLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYE 299
GL G Q+ ++C+EM ++ H Y F + Y+
Sbjct: 231 GLLN---GTGWIYSLLNQNVLMCLEMPFFALSHWYAFRIEDYD 270
>YK31_YEAST (P36142) Hypothetical UPF0206 protein YKR051w
Length = 418
Score = 115 bits (288), Expect = 3e-25
Identities = 92/300 (30%), Positives = 140/300 (46%), Gaps = 47/300 (15%)
Query: 7 LHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVS 66
L+ P V+++I+A++ +S Y + HL Y+ P EQ+ I ++L+VP +S+
Sbjct: 10 LYWPCVYSSIIATI-------ISFYTITRHLLNYRKPYEQRLSIRILLLVPIFSVSCASG 62
Query: 67 LVNPSIS---VDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLL 123
++ P + VD +R+ YE+F +Y F +L LGGE I T L L
Sbjct: 63 IIKPEAAQFYVDP--IREFYEAFVIYTFFTFLTLLLGGERNII------------TVLSL 108
Query: 124 HHHSHDYKGTVNHPFPL-KYFLKPWILTRRF-YQIVKFGIVQYMIIKSFTSIMAVILEAF 181
+H HP PL KP L+ F + VK GI+QY+ K F F
Sbjct: 109 NH------APTRHPIPLIGKICKPIDLSDPFDFLFVKKGILQYVWFKPFY--------CF 154
Query: 182 GVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVF 241
G +KL ++ V N S +W+LY L F+ EL KP KFL K I+F
Sbjct: 155 GTLICSAWKLPKFEIFLNVFYNISVTWSLYSLALFWKCLYPELTPYKPWLKFLCVKLIIF 214
Query: 242 LTWWQGVAIALLYTFGLFKSPIAQGLQSKSS---VQDFIICIEMGIASIVHLYVFPAKPY 298
++WQ + I L G + G Q ++S ++ ++CIEM +I+H FP Y
Sbjct: 215 ASYWQSIIIQGLVVTG----KLGTGNQDRTSGYVYKNGLLCIEMVPFAILHAVAFPWNKY 270
>TTCE_HUMAN (Q96N46) Tetratricopeptide repeat protein 14 (TPR repeat
protein 14)
Length = 770
Score = 32.3 bits (72), Expect = 2.8
Identities = 29/107 (27%), Positives = 46/107 (42%), Gaps = 12/107 (11%)
Query: 364 YIVKDVKFTVHQAVEPVE---KGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPA 420
Y+ K ++ QA + + K I EKL ++ + K+ K RR+ S ++S+
Sbjct: 428 YMQKSLELREKQAEKEEKQKTKKIETSAEKLRKLLKEEKRLKKKRRKSTSSSSVSSAD-- 485
Query: 421 RRVIRGIDDPLLNGSISDSGISRAKKHRRKSGYTSAESGGESSSDQS 467
+ + S S SG R KKH+R +S S SS S
Sbjct: 486 -------ESVSSSSSSSSSGHKRHKKHKRNRSESSRSSRRHSSRASS 525
>COX2_NEUCR (P00411) Cytochrome c oxidase polypeptide II (EC
1.9.3.1)
Length = 250
Score = 32.3 bits (72), Expect = 2.8
Identities = 23/76 (30%), Positives = 35/76 (45%), Gaps = 6/76 (7%)
Query: 186 EGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWW 245
EG +L Y VV+ F W L +++ Y TK ++H K+L +++ L W
Sbjct: 31 EGLVELHDNIMYYLVVILFVVGWILLSIIRNYISTKSPISH-----KYLNHGTLIELIWT 85
Query: 246 QGVAIAL-LYTFGLFK 260
A+ L L F FK
Sbjct: 86 ITPAVILILIAFPSFK 101
>VLF1_NPVOP (O10330) Very late expression factor 1
Length = 374
Score = 32.0 bits (71), Expect = 3.6
Identities = 23/86 (26%), Positives = 39/86 (44%), Gaps = 4/86 (4%)
Query: 388 NEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRG-IDDPLLNGSISDSGISRAKK 446
N + +IS+N KD RR+ D++ + P +IR + L N + ++R
Sbjct: 266 NPTVLQISKNTSTPFKDFRRLLDEAGVEMERPRSNMIRHYLSSNLYNSGVPLQKVARLMN 325
Query: 447 HRRKSG---YTSAESGGESSSDQSYG 469
H + Y + + ESSSD+ G
Sbjct: 326 HESPASTKPYLNKYNFDESSSDEESG 351
>HSLO_SALTY (Q8ZLJ4) 33 kDa chaperonin (Heat shock protein 33
homolog) (HSP33)
Length = 292
Score = 32.0 bits (71), Expect = 3.6
Identities = 17/44 (38%), Positives = 26/44 (58%), Gaps = 1/44 (2%)
Query: 302 GDRHPGSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAK 345
G+R+ G + + GD A C L+ +R + PT+L + T DVD K
Sbjct: 122 GERYQGVVGLEGDTLAAC-LEDYFLRSEQLPTRLFIRTGDVDGK 164
>HSLO_SALTI (Q8Z214) 33 kDa chaperonin (Heat shock protein 33
homolog) (HSP33)
Length = 292
Score = 32.0 bits (71), Expect = 3.6
Identities = 17/44 (38%), Positives = 26/44 (58%), Gaps = 1/44 (2%)
Query: 302 GDRHPGSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAK 345
G+R+ G + + GD A C L+ +R + PT+L + T DVD K
Sbjct: 122 GERYQGVVGLEGDTLAAC-LEDYFLRSEQLPTRLFIRTGDVDGK 164
>COX2_PODAN (P20682) Cytochrome c oxidase polypeptide II (EC
1.9.3.1)
Length = 250
Score = 32.0 bits (71), Expect = 3.6
Identities = 24/76 (31%), Positives = 35/76 (45%), Gaps = 6/76 (7%)
Query: 186 EGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWW 245
EG +L Y VV+ F+ W L +V Y TK ++H K+L +++ L W
Sbjct: 31 EGLNELHDNIMYYLVVILFAVGWILLSIVINYVSTKSPISH-----KYLNHGTLIELIWT 85
Query: 246 QGVAIAL-LYTFGLFK 260
A+ L L F FK
Sbjct: 86 ITPAVILILIAFPSFK 101
>HSLO_ECOLI (P45803) 33 kDa chaperonin (Heat shock protein 33)
(HSP33)
Length = 292
Score = 31.6 bits (70), Expect = 4.8
Identities = 17/44 (38%), Positives = 26/44 (58%), Gaps = 1/44 (2%)
Query: 302 GDRHPGSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAK 345
G+R+ G + + GD A C L+ +R + PT+L + T DVD K
Sbjct: 122 GERYQGVVGLEGDTLAAC-LEDYFMRSEQLPTRLFIRTGDVDGK 164
>APU_THETU (P38536) Amylopullulanase precursor
(Alpha-amylase/pullulanase) (Pullulanase type II)
[Includes: Alpha-amylase (EC 3.2.1.1) (1,4-alpha-D-glucan
glucanohydrolase); Pullulanase (EC 3.2.1.41)
(1,4-alpha-D-glucan glucanohydrolase) (Alpha-dextrin en
Length = 1861
Score = 31.6 bits (70), Expect = 4.8
Identities = 21/63 (33%), Positives = 29/63 (45%), Gaps = 6/63 (9%)
Query: 203 NFSQSWALYCLVQFYTVTKDELAHIKPLAKFL----TFKSIVFLTWWQGVAIALLYTFGL 258
N Q W + FY V D+L IKP A +L T S V LTW +Y + +
Sbjct: 1135 NQGQDWITTSTLSFYVVPSDDL--IKPTAPYLNQPGTESSRVSLTWNPSTDNVGIYDYEI 1192
Query: 259 FKS 261
++S
Sbjct: 1193 YRS 1195
>VIT1_AEDAE (Q16927) Vitellogenin A1 precursor (VG) (PVG1)
[Contains: Vitellin light chain (VL); Vitellin heavy
chain (VH)]
Length = 2148
Score = 31.2 bits (69), Expect = 6.2
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 8/99 (8%)
Query: 377 VEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRGIDDPLLNGSI 436
+ PVE+ + L ++ + ++ +D IK+ + + + ++D + S
Sbjct: 446 ISPVEQ----YKPLLDKVEKRGNRYRRDLNAIKEKKYYEAYKMDQYRLHRLNDTSSDSSS 501
Query: 437 SDSGIS---RAKKHRR-KSGYTSAESGGESSSDQSYGAY 471
SDS S +K+HR S Y+S+ S SSS +Y
Sbjct: 502 SDSSSSSSSESKEHRNGTSSYSSSSSSSSSSSSSESSSY 540
>TGLX_PAPHA (Q8MIC2) Homeobox protein TGIF2LX (TGFB-induced factor
2-like protein, X-linked) (TGF(beta)induced
transcription factor 2-like protein) (TGIF-like on the
X)
Length = 256
Score = 31.2 bits (69), Expect = 6.2
Identities = 20/72 (27%), Positives = 39/72 (53%), Gaps = 8/72 (11%)
Query: 389 EKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRGIDDPLLNGSISDSG--ISRAKK 446
E R+ ++ ++ +KD RR K DS + SPA+ D ++ + +D+G ++ +
Sbjct: 10 ETRSRVEKDSRRVEKDSRRPKKDSPAKTQSPAQ------DTSIMLRNNADTGKVLALPEH 63
Query: 447 HRRKSGYTSAES 458
+++ GY AES
Sbjct: 64 KKKRKGYLPAES 75
>PUR9_SYNY3 (P74741) Bifunctional purine biosynthesis protein purH
[Includes: Phosphoribosylaminoimidazolecarboxamide
formyltransferase (EC 2.1.2.3) (AICAR transformylase);
IMP cyclohydrolase (EC 3.5.4.10) (Inosinicase) (IMP
synthetase) (ATIC)]
Length = 511
Score = 30.8 bits (68), Expect = 8.1
Identities = 26/87 (29%), Positives = 37/87 (41%), Gaps = 8/87 (9%)
Query: 299 ELMGDR----HP---GSISVLGDYSAD-CPLDPDEIRDSERPTKLRLPTTDVDAKSGMTI 350
E++G R HP G I D +D L+ ++IR + P AK G+T+
Sbjct: 58 EILGGRVKTLHPRIHGGILARRDLPSDQADLEANDIRPLDLVVVNLYPFEQTIAKPGVTV 117
Query: 351 RESVRDVVIGGSGYIVKDVKFTVHQAV 377
E+V + IGG I K H V
Sbjct: 118 AEAVEQIDIGGPAMIRATAKNFAHTTV 144
>PUR9_ANASP (Q8YSJ2) Bifunctional purine biosynthesis protein purH
[Includes: Phosphoribosylaminoimidazolecarboxamide
formyltransferase (EC 2.1.2.3) (AICAR transformylase);
IMP cyclohydrolase (EC 3.5.4.10) (Inosinicase) (IMP
synthetase) (ATIC)]
Length = 506
Score = 30.8 bits (68), Expect = 8.1
Identities = 30/109 (27%), Positives = 48/109 (43%), Gaps = 15/109 (13%)
Query: 299 ELMGDR----HP---GSISVLGDYSADCP-LDPDEIRDSERPTKLRLPTTDVDAKSGMTI 350
E++G R HP G I D ++D L+ ++IR + P AK G+T+
Sbjct: 58 EILGGRVKTLHPRIHGGILARRDVASDLTDLENNQIRPIDLVVVNLYPFESTIAKPGVTL 117
Query: 351 RESVRDVVIGGSGYIVKDVKFTVHQAV--EPVEKGITRFNEKLHRISQN 397
E+V + IGG + K H V +P + ++E L + QN
Sbjct: 118 AEAVEQIDIGGPAMLRASSKNFAHLTVLCDPAQ-----YDEYLQELRQN 161
>FRDD_VIBVU (Q8DCX4) Fumarate reductase subunit D
Length = 125
Score = 30.8 bits (68), Expect = 8.1
Identities = 14/52 (26%), Positives = 29/52 (54%), Gaps = 2/52 (3%)
Query: 15 TIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVS 66
+I+ ++F++ TLAL M+ + + + KF GV+ + CY++ +S
Sbjct: 66 SIIGALFIIATLALPMWHAMHRV--HHGMHDLKFHTGVVGKIACYAVAGLIS 115
>CLI5_BOVIN (P35526) Chloride intracellular channel protein 5
(Chlorine channel protein p64)
Length = 437
Score = 30.8 bits (68), Expect = 8.1
Identities = 18/44 (40%), Positives = 21/44 (46%), Gaps = 2/44 (4%)
Query: 291 YVFPAKPYELMGDRHPGSISVLGDYSADCPLDP--DEIRDSERP 332
YV P +PY D HPG + D S + L P D DSE P
Sbjct: 81 YVLPDEPYSKAQDPHPGEPTADEDISLEELLSPTKDHQSDSEEP 124
>APU_THESA (P36905) Amylopullulanase precursor
(Alpha-amylase/pullulanase) [Includes: Alpha-amylase (EC
3.2.1.1) (1,4-alpha-D-glucan glucanohydrolase);
Pullulanase (EC 3.2.1.41) (1,4-alpha-D-glucan
glucanohydrolase) (Alpha-dextrin endo-1,6-alpha-glucosida
Length = 1279
Score = 30.8 bits (68), Expect = 8.1
Identities = 20/63 (31%), Positives = 28/63 (43%), Gaps = 6/63 (9%)
Query: 203 NFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSI----VFLTWWQGVAIALLYTFGL 258
N QSW + FY V D+L IKP A L + V LTW +Y + +
Sbjct: 1135 NQGQSWTTTDTLSFYVVPSDDL--IKPTAPILNQPGVESSRVSLTWSPSTDNVGIYNYEI 1192
Query: 259 FKS 261
++S
Sbjct: 1193 YRS 1195
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,686,513
Number of Sequences: 164201
Number of extensions: 2382416
Number of successful extensions: 5490
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 5475
Number of HSP's gapped (non-prelim): 20
length of query: 482
length of database: 59,974,054
effective HSP length: 114
effective length of query: 368
effective length of database: 41,255,140
effective search space: 15181891520
effective search space used: 15181891520
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0117b.4


