

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0114.8
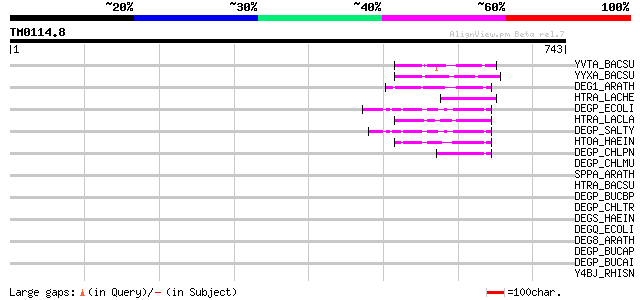
(743 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YVTA_BACSU (Q9R9I1) Probable serine protease yvtA (EC 3.4.21.-) 53 3e-06
YYXA_BACSU (P39668) Hypothetical serine protease yyxA (EC 3.4.21.-) 51 1e-05
DEG1_ARATH (O22609) Protease Do-like 1, chloroplast precursor (E... 50 2e-05
HTRA_LACHE (Q9Z4H7) Serine protease do-like htrA (EC 3.4.21.-) 49 6e-05
DEGP_ECOLI (P09376) Protease do precursor (EC 3.4.21.-) 49 6e-05
HTRA_LACLA (Q9LA06) Serine protease do-like htrA (EC 3.4.21.-) (... 48 1e-04
DEGP_SALTY (P26982) Protease do precursor (EC 3.4.21.-) 47 2e-04
HTOA_HAEIN (P45129) Probable periplasmic serine protease do/hhoA... 46 4e-04
DEGP_CHLPN (Q9Z6T0) Probable serine protease do-like precursor (... 45 5e-04
DEGP_CHLMU (Q9PL97) Probable serine protease do-like precursor (... 45 0.001
SPPA_ARATH (Q9SEL7) Protease sppA, chloroplast precursor (EC 3.4... 44 0.001
HTRA_BACSU (O34358) Probable serine protease do-like htrA (EC 3.... 44 0.001
DEGP_BUCBP (Q89AP5) Probable serine protease do-like precursor (... 43 0.003
DEGP_CHLTR (P18584) Probable serine protease do-like precursor (... 43 0.003
DEGS_HAEIN (P44947) Protease degS precursor (EC 3.4.21.-) 42 0.005
DEGQ_ECOLI (P39099) Protease degQ precursor (EC 3.4.21.-) 42 0.006
DEG8_ARATH (Q9LU10) Protease Do-like 8, chloroplast precursor (E... 41 0.013
DEGP_BUCAP (O85291) Probable serine protease do-like precursor (... 40 0.017
DEGP_BUCAI (P57322) Probable serine protease do-like precursor (... 40 0.029
Y4BJ_RHISN (P55377) Hypothetical 67.9 kDa protein Y4BJ 39 0.038
>YVTA_BACSU (Q9R9I1) Probable serine protease yvtA (EC 3.4.21.-)
Length = 458
Score = 52.8 bits (125), Expect = 3e-06
Identities = 43/139 (30%), Positives = 69/139 (48%), Gaps = 21/139 (15%)
Query: 516 DVALLQIESIPDNLVPMVMNFSRPSTGSKAYVIGHGLFGPKCGFFPSVCSGVIA---KVV 572
D+A+L+I V + S+ TG K IG+ L G + F +V G+I+ + +
Sbjct: 217 DLAVLEISGKNVKKVASFGDSSQLRTGEKVIAIGNPL-GQQ--FSGTVTQGIISGLNRTI 273
Query: 573 EAKTTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSG 632
+ TTQ + +L+T AA++PG SGG +IN+ G +IG+ + SG
Sbjct: 274 DVDTTQGTVEMN------------VLQTDAAINPGNSGGPLINASGQVIGINSLKVSESG 321
Query: 633 GAVIPHLNFSIPSAALAPI 651
+ L F+IPS + PI
Sbjct: 322 ---VESLGFAIPSNDVEPI 337
Score = 32.7 bits (73), Expect = 3.6
Identities = 40/164 (24%), Positives = 75/164 (45%), Gaps = 24/164 (14%)
Query: 228 SLTRMAILSVS-LSLKDSLDSSVCSMIKRGDFLLAIGSPF----------GVLSPTHFFN 276
++T +A+L +S ++K S ++ G+ ++AIG+P G++S +
Sbjct: 214 TITDLAVLEISGKNVKKVASFGDSSQLRTGEKVIAIGNPLGQQFSGTVTQGIISGLN--R 271
Query: 277 SYPPNSSDG----SLLMADIRTLPGMEGSPVFSEHACLTGVLIRPLRQKTSGVET-QLVI 331
+ +++ G ++L D PG G P+ + + G I L+ SGVE+ I
Sbjct: 272 TIDVDTTQGTVEMNVLQTDAAINPGNSGGPLINASGQVIG--INSLKVSESGVESLGFAI 329
Query: 332 PWEAIVTASSGLLRKWGK--GPFVGTYKLDV-PASNTHEQLNLG 372
P + LL+ GK PF+G +D+ T+++ LG
Sbjct: 330 PSNDVEPIVDQLLQN-GKVDRPFLGVQMIDMSQVPETYQENTLG 372
>YYXA_BACSU (P39668) Hypothetical serine protease yyxA (EC 3.4.21.-)
Length = 400
Score = 50.8 bits (120), Expect = 1e-05
Identities = 41/141 (29%), Positives = 66/141 (46%), Gaps = 12/141 (8%)
Query: 516 DVALLQIESIPDNLVPMVMNFSRPSTGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEAK 575
D+A+L+++S V N + +G IG+ L F SV GVI+ A
Sbjct: 159 DLAVLRVKSDKIKAVADFGNSDKVKSGEPVIAIGNPL---GLEFAGSVTQGVISGTERAI 215
Query: 576 TTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAV 635
S Q + +L+T AA++PG SGGA++N DG +IG+ N+ +
Sbjct: 216 PVDSNGDGQPDWN------AEVLQTDAAINPGNSGGALLNMDGKVIGI---NSMKIAESA 266
Query: 636 IPHLNFSIPSAALAPIFKFAE 656
+ + SIPS + P+ + E
Sbjct: 267 VEGIGLSIPSKLVIPVIEDLE 287
>DEG1_ARATH (O22609) Protease Do-like 1, chloroplast precursor (EC
3.4.21.-)
Length = 439
Score = 50.1 bits (118), Expect = 2e-05
Identities = 44/142 (30%), Positives = 71/142 (49%), Gaps = 19/142 (13%)
Query: 504 DAKVVYICKGPWDVALLQIESIPDNLVPMVMNFSRPS-TGSKAYVIGHGLFGPKCGFFPS 562
DAKVV + DVA+L+I++ + L P+ + S G K + IG+ FG
Sbjct: 192 DAKVVGFDQDK-DVAVLRIDAPKNKLRPIPVGVSADLLVGQKVFAIGNP-FGLDHTLTTG 249
Query: 563 VCSGVIAKVVEAKTTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIG 622
V SG+ ++ A T + Q + ++T AA++PG SGG +++S G +IG
Sbjct: 250 VISGLRREISSAATGRPIQDV--------------IQTDAAINPGNSGGPLLDSSGTLIG 295
Query: 623 LVTSNARHSGGAVIPHLNFSIP 644
+ T+ SG + + FSIP
Sbjct: 296 INTAIYSPSGAS--SGVGFSIP 315
>HTRA_LACHE (Q9Z4H7) Serine protease do-like htrA (EC 3.4.21.-)
Length = 413
Score = 48.5 bits (114), Expect = 6e-05
Identities = 29/77 (37%), Positives = 46/77 (59%), Gaps = 2/77 (2%)
Query: 577 TQSYQSIQAEHMHTN-GNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSN-ARHSGGA 634
TQ S A + T+ GN +++T AA++PG SGGA++NS G +IG+ + A+ S G
Sbjct: 225 TQGIISAPARTISTSSGNQQTVIQTDAAINPGNSGGALVNSAGQVIGINSMKLAQSSDGT 284
Query: 635 VIPHLNFSIPSAALAPI 651
+ + F+IPS + I
Sbjct: 285 SVEGMAFAIPSNEVVTI 301
>DEGP_ECOLI (P09376) Protease do precursor (EC 3.4.21.-)
Length = 474
Score = 48.5 bits (114), Expect = 6e-05
Identities = 52/177 (29%), Positives = 88/177 (49%), Gaps = 30/177 (16%)
Query: 473 DEHGGYKLNPSY--DNHRNIRIRLDHIKPWVWCDAKVVYICKGPW-DVALLQIESIPDNL 529
D GY + ++ DN I+++L + + DAK+V K P D+AL+QI++ P NL
Sbjct: 120 DADKGYVVTNNHVVDNATVIKVQLSDGRKF---DAKMVG--KDPRSDIALIQIQN-PKNL 173
Query: 530 VPMVMNFSRP-STGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEAKTTQSYQSIQAEHM 588
+ M S G IG+ G +V SG+++ + + + AE
Sbjct: 174 TAIKMADSDALRVGDYTVAIGNPF-----GLGETVTSGIVSALGRS-------GLNAE-- 219
Query: 589 HTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAVIPHLNFSIPS 645
N+ ++T AA++ G SGGA++N +G +IG+ T+ GG + + F+IPS
Sbjct: 220 ----NYENFIQTDAAINRGNSGGALVNLNGELIGINTAILAPDGGNI--GIGFAIPS 270
>HTRA_LACLA (Q9LA06) Serine protease do-like htrA (EC 3.4.21.-)
(HtrALl)
Length = 408
Score = 47.8 bits (112), Expect = 1e-04
Identities = 39/132 (29%), Positives = 62/132 (46%), Gaps = 13/132 (9%)
Query: 516 DVALLQIESIPDNLVPMVMNFSRPSTGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEAK 575
D+A+L+I S V + S+ + G A +G L F + G+++
Sbjct: 157 DLAVLKISSEHVKDVATFADSSKLTIGEPAIAVGSPLGSQ---FANTATEGILSA----- 208
Query: 576 TTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTS--NARHSGG 633
T++ Q TN N ++T AA++PG SGGA+IN +G +IG+ S G
Sbjct: 209 TSRQVTLTQENGQTTNIN---AIQTDAAINPGNSGGALINIEGQVIGITQSKITTTEDGS 265
Query: 634 AVIPHLNFSIPS 645
+ L F+IPS
Sbjct: 266 TSVEGLGFAIPS 277
>DEGP_SALTY (P26982) Protease do precursor (EC 3.4.21.-)
Length = 475
Score = 46.6 bits (109), Expect = 2e-04
Identities = 49/167 (29%), Positives = 82/167 (48%), Gaps = 28/167 (16%)
Query: 481 NPSYDNHRNIRIRLDHIKPWVWCDAKVVYICKGPW-DVALLQIESIPDNLVPMVMNFSRP 539
N DN I+++L + + DAKVV K P D+AL+QI++ P NL + + S
Sbjct: 131 NHVVDNASVIKVQLSDGRKF---DAKVVG--KDPRSDIALIQIQN-PKNLTAIKLADSDA 184
Query: 540 -STGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEAKTTQSYQSIQAEHMHTNGNFPAML 598
G IG+ G +V SG+++ + + + E N+ +
Sbjct: 185 LRVGDYTVAIGNPF-----GLGETVTSGIVSALGRS-------GLNVE------NYENFI 226
Query: 599 ETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAVIPHLNFSIPS 645
+T AA++ G SGGA++N +G +IG+ T+ GG + + F+IPS
Sbjct: 227 QTDAAINRGNSGGALVNLNGELIGINTAILAPDGGNI--GIGFAIPS 271
>HTOA_HAEIN (P45129) Probable periplasmic serine protease
do/hhoA-like precursor (EC 3.4.21.-)
Length = 466
Score = 45.8 bits (107), Expect = 4e-04
Identities = 39/131 (29%), Positives = 64/131 (48%), Gaps = 21/131 (16%)
Query: 516 DVALLQIESIPDNLVPMVMNFS-RPSTGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEA 574
D+AL+Q+E P NL + S + G IG+ G +V SG+++ + +
Sbjct: 150 DIALVQLEK-PSNLTEIKFADSDKLRVGDFTVAIGNPF-----GLGQTVTSGIVSALGRS 203
Query: 575 KTTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGA 634
+ S G + ++T AAV+ G SGGA++N +G +IG+ T+ SGG
Sbjct: 204 TGSDS------------GTYENYIQTDAAVNRGNSGGALVNLNGELIGINTAIISPSGGN 251
Query: 635 VIPHLNFSIPS 645
+ F+IPS
Sbjct: 252 A--GIAFAIPS 260
>DEGP_CHLPN (Q9Z6T0) Probable serine protease do-like precursor (EC
3.4.21.-)
Length = 488
Score = 45.4 bits (106), Expect = 5e-04
Identities = 25/74 (33%), Positives = 42/74 (55%), Gaps = 2/74 (2%)
Query: 572 VEAKTTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHS 631
++A T S + + +F ++T AA++PG SGG ++N DG +IG+ T+ S
Sbjct: 201 LQATVTVGVISAKGRNQLHIADFEDFIQTDAAINPGNSGGPLLNIDGQVIGVNTAIVSGS 260
Query: 632 GGAVIPHLNFSIPS 645
GG + + F+IPS
Sbjct: 261 GGYI--GIGFAIPS 272
>DEGP_CHLMU (Q9PL97) Probable serine protease do-like precursor (EC
3.4.21.-)
Length = 497
Score = 44.7 bits (104), Expect = 0.001
Identities = 22/53 (41%), Positives = 35/53 (65%), Gaps = 2/53 (3%)
Query: 593 NFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAVIPHLNFSIPS 645
+F ++T AA++PG SGG ++N DG +IG+ T+ SGG + + F+IPS
Sbjct: 231 DFEDFIQTDAAINPGNSGGPLLNIDGQVIGVNTAIVSGSGGYI--GIGFAIPS 281
>SPPA_ARATH (Q9SEL7) Protease sppA, chloroplast precursor (EC
3.4.21.-)
Length = 321
Score = 44.3 bits (103), Expect = 0.001
Identities = 34/130 (26%), Positives = 63/130 (48%), Gaps = 17/130 (13%)
Query: 516 DVALLQIESIPDNLVPMVMNFSRP-STGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEA 574
D+A+L+IE+ L P+V+ S G + IG+ G+ ++ GV++ +
Sbjct: 186 DLAVLKIETEGRELNPVVLGTSNDLRVGQSCFAIGNPY-----GYENTLTIGVVSGLGRE 240
Query: 575 KTTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGA 634
+ + +SI ++T A ++ G SGG +++S GH IG+ T+ G
Sbjct: 241 IPSPNGKSISEA-----------IQTDADINSGNSGGPLLDSYGHTIGVNTATFTRKGSG 289
Query: 635 VIPHLNFSIP 644
+ +NF+IP
Sbjct: 290 MSSGVNFAIP 299
>HTRA_BACSU (O34358) Probable serine protease do-like htrA (EC
3.4.21.-)
Length = 449
Score = 44.3 bits (103), Expect = 0.001
Identities = 42/142 (29%), Positives = 66/142 (45%), Gaps = 27/142 (19%)
Query: 516 DVALLQIESIPDNLVPMVMNFSRPS---TGSKAYVIGHGLFGPKCGFFPSVCSGVIAKV- 571
D+A+LQI D+ V V NF S TG IG L +V G+++ V
Sbjct: 209 DLAVLQIS---DDHVTKVANFGDSSDLRTGETVIAIGDPLGKD---LSRTVTQGIVSGVD 262
Query: 572 --VEAKTTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNAR 629
V T+ SI +++T AA++PG SGG ++N+DG ++G+ N+
Sbjct: 263 RTVSMSTSAGETSIN------------VIQTDAAINPGNSGGPLLNTDGKIVGI---NSM 307
Query: 630 HSGGAVIPHLNFSIPSAALAPI 651
+ + F+IPS + PI
Sbjct: 308 KISEDDVEGIGFAIPSNDVKPI 329
>DEGP_BUCBP (Q89AP5) Probable serine protease do-like precursor (EC
3.4.21.-)
Length = 465
Score = 43.1 bits (100), Expect = 0.003
Identities = 43/179 (24%), Positives = 79/179 (44%), Gaps = 36/179 (20%)
Query: 473 DEHGGYKLNPSY--DNHRNIRIRLDHIKPWVWCDAKVVYICKGP-WDVALLQIESIPDNL 529
D GY + S+ D I+++L + C + V I K +D+A+++++ + +
Sbjct: 108 DSKNGYIVTNSHVVDRANKIQVQLSN-----GCKHEAVVIGKDARFDIAIIKLKKVKNLH 162
Query: 530 VPMVMNFSRPSTGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEAKTTQSYQSIQAEHMH 589
+ N G IG+ G +V SG+I+ +H
Sbjct: 163 EIKMSNSDILKVGDYVIAIGNPY-----GLGETVTSGIISA-----------------LH 200
Query: 590 TNG----NFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAVIPHLNFSIP 644
+G N+ ++T AA++ G SGGA++N G +IG+ T+ GG + + F+IP
Sbjct: 201 RSGLNIENYENFIQTDAAINRGNSGGALVNLKGELIGINTAILTPDGGNI--GIGFAIP 257
>DEGP_CHLTR (P18584) Probable serine protease do-like precursor (EC
3.4.21.-) (59 kDa immunogenic protein) (SK59)
Length = 497
Score = 42.7 bits (99), Expect = 0.003
Identities = 21/53 (39%), Positives = 35/53 (65%), Gaps = 2/53 (3%)
Query: 593 NFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAVIPHLNFSIPS 645
+F ++T AA++PG SGG ++N +G +IG+ T+ SGG + + F+IPS
Sbjct: 231 DFEDFIQTDAAINPGNSGGPLLNINGQVIGVNTAIVSGSGGYI--GIGFAIPS 281
>DEGS_HAEIN (P44947) Protease degS precursor (EC 3.4.21.-)
Length = 340
Score = 42.4 bits (98), Expect = 0.005
Identities = 37/130 (28%), Positives = 61/130 (46%), Gaps = 20/130 (15%)
Query: 516 DVALLQIESIPDNLVPMVMNFSRPS-TGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEA 574
D+A+L+I + DNL + N +R + G IG+ SV G+I+ +
Sbjct: 122 DLAVLKIRA--DNLSTIPQNSARQAHVGDVVLAIGNPY-----NLGQSVSQGIISAIGRN 174
Query: 575 KTTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGA 634
S NF ++T A+++ G SGGA+INS G ++G+ T + +
Sbjct: 175 AVGDSVG---------RQNF---IQTDASINRGNSGGALINSAGELVGISTLSIGKTANE 222
Query: 635 VIPHLNFSIP 644
+ LNF+IP
Sbjct: 223 IAEGLNFAIP 232
>DEGQ_ECOLI (P39099) Protease degQ precursor (EC 3.4.21.-)
Length = 455
Score = 42.0 bits (97), Expect = 0.006
Identities = 55/217 (25%), Positives = 104/217 (47%), Gaps = 38/217 (17%)
Query: 441 LEGTTSLGNKIESDQISQTLHSKVPVH--YPFAADEHG-------GYKLNPSY--DNHRN 489
+EGT S G KI ++ + +P PF G GY L ++ + +
Sbjct: 58 VEGTASQGQKIP-EEFKKFFGDDLPDQPAQPFEGLGSGVIINASKGYVLTNNHVINQAQK 116
Query: 490 IRIRLDHIKPWVWCDAKVVYICKGPWDVALLQIESIPDNLVPM-VMNFSRPSTGSKAYVI 548
I I+L+ + + DAK++ D+ALLQI++ P L + + + + G A +
Sbjct: 117 ISIQLNDGREF---DAKLIG-SDDQSDIALLQIQN-PSKLTQIAIADSDKLRVGDFAVAV 171
Query: 549 GHGLFGPKCGFFPSVCSGVIAKVVEAKTTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGA 608
G+ G + SG+++ + ++ + + ++ NF ++T A+++ G
Sbjct: 172 GNPF-----GLGQTATSGIVSAL--GRSGLNLEGLE--------NF---IQTDASINRGN 213
Query: 609 SGGAVINSDGHMIGLVTSNARHSGGAVIPHLNFSIPS 645
SGGA++N +G +IG+ T+ GG+V + F+IPS
Sbjct: 214 SGGALLNLNGELIGINTAILAPGGGSV--GIGFAIPS 248
>DEG8_ARATH (Q9LU10) Protease Do-like 8, chloroplast precursor (EC
3.4.21.-)
Length = 448
Score = 40.8 bits (94), Expect = 0.013
Identities = 61/289 (21%), Positives = 117/289 (40%), Gaps = 83/289 (28%)
Query: 364 NTHEQLNLGSSSLLPIEKAMASVCLVTIGDGVWASGILLNSQGLVLTNAHLLEPWRFGKT 423
NT+ +N+ +L P K M V + G+G SG++ + QG ++TN H++
Sbjct: 126 NTYSVVNIFDVTLRPQLK-MTGVVEIPEGNG---SGVVWDGQGYIVTNYHVI-------- 173
Query: 424 HVSGGGNGTNSKTLPFMLEGTTSLGNKIESDQISQTLHSKVPVHYPFAADEHGGYKLNPS 483
GN + P + G ++ + SD + + K+
Sbjct: 174 -----GNALSRNPSPGDVVGRVNI---LASDGVQKNFEGKL------------------- 206
Query: 484 YDNHRNIRIRLDHIKPWVWCDAKVVYICKGPWDVALLQIESIPDNLVPMVMNFSRP-STG 542
+ D K D+A+L++++ L P+ + S G
Sbjct: 207 --------VGADRAK-----------------DLAVLKVDAPETLLKPIKVGQSNSLKVG 241
Query: 543 SKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEAKTTQSYQSIQAEHMHTNGNFPAMLETTA 602
+ IG+ GF ++ GVI+ + +Q+ +I ++T A
Sbjct: 242 QQCLAIGNPF-----GFDHTLTVGVISGLNRDIFSQTGVTIGGG-----------IQTDA 285
Query: 603 AVHPGASGGAVINSDGHMIGLVTSNARHSGGAVIPHLNFSIPSAALAPI 651
A++PG SGG +++S G++IG+ T+ +G + + F+IPS+ + I
Sbjct: 286 AINPGNSGGPLLDSKGNLIGINTAIFTQTGTSA--GVGFAIPSSTVLKI 332
>DEGP_BUCAP (O85291) Probable serine protease do-like precursor (EC
3.4.21.-)
Length = 478
Score = 40.4 bits (93), Expect = 0.017
Identities = 34/129 (26%), Positives = 61/129 (46%), Gaps = 20/129 (15%)
Query: 516 DVALLQIESIPDNLVPMVMNFSRPSTGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEAK 575
D+AL+Q+++ + + + G IG+ G +V SG+I+ + +
Sbjct: 163 DIALIQLKNAKNLSAIKIADSDTLRVGDYTVAIGNPY-----GLGETVTSGIISALGRS- 216
Query: 576 TTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAV 635
+ EH NF ++T AA++ G SGGA++N G +IG+ T+ GG +
Sbjct: 217 ------GLNIEHYE---NF---IQTDAAINRGNSGGALVNLKGELIGINTAILAPDGGNI 264
Query: 636 IPHLNFSIP 644
+ F+IP
Sbjct: 265 --GIGFAIP 271
>DEGP_BUCAI (P57322) Probable serine protease do-like precursor (EC
3.4.21.-)
Length = 478
Score = 39.7 bits (91), Expect = 0.029
Identities = 34/129 (26%), Positives = 61/129 (46%), Gaps = 20/129 (15%)
Query: 516 DVALLQIESIPDNLVPMVMNFSRPSTGSKAYVIGHGLFGPKCGFFPSVCSGVIAKVVEAK 575
D+AL+Q+++ + + + G IG+ G +V SG+I+ + +
Sbjct: 163 DIALIQLKNANNLSEIKIADSDNLRVGDYTVAIGNPY-----GLGETVTSGIISALGRS- 216
Query: 576 TTQSYQSIQAEHMHTNGNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSNARHSGGAV 635
+ EH NF ++T AA++ G SGGA++N G +IG+ T+ GG +
Sbjct: 217 ------GLNIEHYE---NF---IQTDAAINRGNSGGALVNLKGELIGINTAILAPDGGNI 264
Query: 636 IPHLNFSIP 644
+ F+IP
Sbjct: 265 --GIGFAIP 271
>Y4BJ_RHISN (P55377) Hypothetical 67.9 kDa protein Y4BJ
Length = 630
Score = 39.3 bits (90), Expect = 0.038
Identities = 23/62 (37%), Positives = 37/62 (59%), Gaps = 5/62 (8%)
Query: 592 GNFPAMLETTAAVHPGASGGAVINSDGHMIGLVTSN-----ARHSGGAVIPHLNFSIPSA 646
GN ++ +A V PG SGG +I+S G++IG+VTS A G + ++NF++ A
Sbjct: 530 GNDTRFIQISAPVQPGNSGGPLIDSYGNVIGVVTSKLDALAALAVTGDIPQNVNFALRGA 589
Query: 647 AL 648
+L
Sbjct: 590 SL 591
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.135 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 89,657,226
Number of Sequences: 164201
Number of extensions: 3898035
Number of successful extensions: 8440
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 8370
Number of HSP's gapped (non-prelim): 83
length of query: 743
length of database: 59,974,054
effective HSP length: 118
effective length of query: 625
effective length of database: 40,598,336
effective search space: 25373960000
effective search space used: 25373960000
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0114.8


