

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
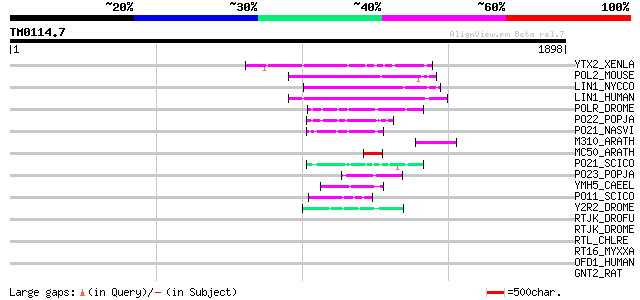
Query= TM0114.7
(1898 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 179 8e-44
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 169 5e-41
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 166 6e-40
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 164 2e-39
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 116 7e-25
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 82 1e-14
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 82 2e-14
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 81 2e-14
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 71 3e-11
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 66 8e-10
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 65 1e-09
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 65 2e-09
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 48 3e-04
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 46 9e-04
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 43 0.010
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 42 0.012
RTL_CHLRE (P11660) Reverse transcriptase-like protein 39 0.18
RT16_MYXXA (P23072) RNA-directed DNA polymerase from retron Mx16... 38 0.31
OFD1_HUMAN (O75665) Oral-facial-digital syndrome 1 protein (Prot... 36 0.89
GNT2_RAT (Q09326) Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acety... 35 1.5
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein (ORF
2)
Length = 1308
Score = 179 bits (453), Expect = 8e-44
Identities = 170/665 (25%), Positives = 302/665 (44%), Gaps = 58/665 (8%)
Query: 808 SDHCPILLQCGSPQTLAHNRPFRFLAAWADHPDFAAVVDNAWRSGEECITSKLERVREAS 867
SDH + L+ +L + F + + FA V + WR G + + +
Sbjct: 228 SDHNCVSLRMSIAPSLPKAAYWHFNNSLLEDEGFAKSVRDTWR-GWRAFQDEFATLNQWW 286
Query: 868 TV-----------FNKEVFGNIFRRKRHVEARLRGVQRELDRRVTS--DMVLFEAELQRE 914
V + K V G +R +EA L G +L++R++ D L L+R+
Sbjct: 287 DVGKVHLKLLCQEYTKSVSG---QRNAEIEA-LNGEVLDLEQRLSGSEDQALQCEYLERK 342
Query: 915 --YRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLEDGTWCGDE 972
RN+ +++ + ++R + DR + +F+ + + ++ L EDGT D
Sbjct: 343 EALRNMEQRQARGAFVRSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDP 402
Query: 973 EVLKRVVHGFFVNLFTG--ITPLASPRVTHDLFPSLSVEAKSLLLQPVRKDEVRHALMSM 1030
E ++ F+ NLF+ I+P A + L P +S K L P+ DE+ AL M
Sbjct: 403 EAIRDRARSFYQNLFSPDPISPDACEELWDGL-PVVSERRKERLETPITLDELSQALRLM 461
Query: 1031 NSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDILVVLIPKVDQPST 1090
+PG DG F++ +W+ +G ++ EAF++G++ + ++ L+PK
Sbjct: 462 PHNKSPGLDGLTIEFFQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRL 521
Query: 1091 VRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHM 1150
+++ RP+SL + +K++ K + RL+ L ++I P QS +PGR DN FL ++++H
Sbjct: 522 IKNWRPVSLLSTDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFA 581
Query: 1151 SKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQ--VSILW 1208
++ + +D EKA+D V +L TL+ Y F + + + S++ V I W
Sbjct: 582 RRTGL--SLAFLSLDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINW 639
Query: 1209 NGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHL 1268
+ AF GRG+RQG P+S L+ L +E L++ + + L + +
Sbjct: 640 SLTAPLAF--GRGVRQGCPLSGQLYSLAIEPFLCLLRKRL-----TGLVLKEPDMRVVLS 692
Query: 1269 FFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVA 1328
+ADDV+L + + + +++ +S +IN SKS S +++ S+ V
Sbjct: 693 AYADDVILVAQ-DLVDLERAQECQEVYAAASSARINWSKS--------SGLLEGSLKVDF 743
Query: 1329 PIPFVHDLG------KYLGFPLKGGRIH-RNRFNFLLESIQRKLGSWR--ANMLSLAGRV 1379
P D+ KYLG L F L E + +LG W+ A +LS+ GR
Sbjct: 744 LPPAFRDISWESKIIKYLGVYLSAEEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRGRA 803
Query: 1380 CLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPKDR 1439
+ ++A+ Y + + + KI + + F+W GK H V+ + P
Sbjct: 804 LVINQLVASQIWYRLICLSPTQEFIAKIQRRLLDFLWIGK------HWVSAGVSSLPLKE 857
Query: 1440 GGLGV 1444
GG GV
Sbjct: 858 GGQGV 862
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 169 bits (429), Expect = 5e-41
Identities = 140/523 (26%), Positives = 238/523 (44%), Gaps = 34/523 (6%)
Query: 953 RRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLF-TGITPLASPRVTHDLF--PSLSVE 1009
R K +++++ E G D E ++ + F+ L+ T + L D + P L+ +
Sbjct: 411 RDKILINKIRNEKGDITTDPEEIQNTIRSFYKRLYSTKLENLDEMDKFLDRYQVPKLNQD 470
Query: 1010 AKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGK 1069
L P+ E+ + S+ + +PG DGF FY+ + + L + + EG
Sbjct: 471 QVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGT 530
Query: 1070 VNENLLDILVVLIPKVDQ-PSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQS 1128
+ + + + LIPK + P+ + + RPISL N+ K++ K+L NR++ ++ +I P Q
Sbjct: 531 LPNSFYEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQV 590
Query: 1129 SFLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFP 1188
F+PG N + VIH+++K K M+ +D EKA+D + F+ + LE G
Sbjct: 591 GFIPGMQGWFNIRKSINVIHYINKLKDKNHMI-ISLDAEKAFDKIQHPFMIKVLERSGIQ 649
Query: 1189 EVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLV 1248
+N+I + + +I NG +L A G RQG P+SPYLF + +E VL +++
Sbjct: 650 GPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLE---VLARAIR 706
Query: 1249 DKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKS 1308
+ + K I++ K IS L ADD++++ L + + F + G KIN +KS
Sbjct: 707 QQKEIKGIQIGKEEVKISLL--ADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKS 764
Query: 1309 KAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPL--KGGRIHRNRFNFLLESIQRKLG 1366
A + + K+ I P V + KYLG L + ++ F L + I+ L
Sbjct: 765 MAFLYTKNKQAEKE-IRETTPFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDLR 823
Query: 1367 SWRANMLSLAGRVCLAKSVIAAMPTYTM---------QVFNMPRGVTCKINQLIRSFIWS 1417
W+ S GR+ + K I Y Q FN G CK F+W+
Sbjct: 824 RWKDLPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICK-------FVWN 876
Query: 1418 GKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIW 1460
K L + + GG+ + D +L A++ K W
Sbjct: 877 NKKPRIAKSL-----LKDKRTSGGITMPDLKLYYRAIVIKTAW 914
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 166 bits (420), Expect = 6e-40
Identities = 128/478 (26%), Positives = 222/478 (45%), Gaps = 22/478 (4%)
Query: 1004 PSLSVEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKE 1063
P LS + +L +P+ E+ + ++ +PG DGF FY+ + + L ++ +
Sbjct: 438 PRLSQKEVEMLNRPISSSEIASTIQNLPKKKSPGPDGFTSEFYQTFKEELVPILLNLFQN 497
Query: 1064 AFQEGKVNENLLDILVVLIPKVDQ-PSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKL 1122
+EG + + + LIPK + P+ + RPISL N+ K++ K+L NR++ ++K+
Sbjct: 498 IEKEGILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKI 557
Query: 1123 IGPMQSSFLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETL 1182
I Q F+PG N + VI H++K K M+ ID EKA+D++ F+ TL
Sbjct: 558 IHHDQVGFIPGSQGWFNIRKSINVIQHINKLKNKDHMI-LSIDAEKAFDNIQHPFMIRTL 616
Query: 1183 ELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSV 1242
+ G + LI + + +I+ NG +L +F G RQG P+SP LF + ME L++
Sbjct: 617 KKIGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVMEVLAI 676
Query: 1243 LIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLK 1302
I+ ++ K I + +S FADD++++ E + L ++ + + SG K
Sbjct: 677 AIR---EEKAIKGIHIGSEEIKLS--LFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYK 731
Query: 1303 INLSKSKAIV---SKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRFNFLLE 1359
IN KS A + + + VKDSI + LG YL +K +++ + L +
Sbjct: 732 INTHKSVAFIYTNNNQAEKTVKDSIPFTVVPKKMKYLGVYLTKDVKD--LYKENYETLRK 789
Query: 1360 SIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQV--FNMPRGVTCKINQLIRSFIWS 1417
I + W+ S GR+ + K I Y P + ++I FIW+
Sbjct: 790 EIAEDVNKWKNIPCSWLGRINIVKMSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIWN 849
Query: 1418 GKGGGHGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIWCLMHKPNK--LWVRV 1473
K L ++ GG+ + D L +++ K W HK + +W R+
Sbjct: 850 QKKPQIAKTL-----LSNKNKAGGITLPDLRLYYKSIVIKTAW-YWHKNREVDVWNRI 901
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 164 bits (415), Expect = 2e-39
Identities = 133/556 (23%), Positives = 248/556 (43%), Gaps = 23/556 (4%)
Query: 952 RRRKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFTG----ITPLASPRVTHDLFPSLS 1007
+R KN++ +K + G D ++ + ++ +L+ + + T+ L P L+
Sbjct: 383 KREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLYANKLENLEEMDKFLDTYTL-PRLN 441
Query: 1008 VEAKSLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQE 1067
E L +P+ E+ + S+ + +PG +GF FY++Y + L + + +E
Sbjct: 442 QEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQSIEKE 501
Query: 1068 GKVNENLLDILVVLIPKVDQPSTVRD-LRPISLCNVAFKVITKVLVNRLRPFLRKLIGPM 1126
G + + + ++LIPK + +T ++ RPISL N+ K++ K+L N+++ ++KLI
Sbjct: 502 GILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIHHD 561
Query: 1127 QSSFLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYG 1186
Q F+P N + +I H++++ M+ ID EKA+D + F+ + L G
Sbjct: 562 QVGFIPAMQGWFNIRKSINIIQHINRTKDTNHMI-ISIDAEKAFDKIQQPFMLKPLNKLG 620
Query: 1187 FPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQS 1246
+ +I + +I+ NG +L A G RQG P+SP L + +E VL ++
Sbjct: 621 IDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSPLLPNIVLE---VLARA 677
Query: 1247 LVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLS 1306
+ + + K I+L K +S FADD++++ E + L + F SG KIN+
Sbjct: 678 IRQEKEIKGIQLGKEEVKLS--LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQ 735
Query: 1307 KSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGG--RIHRNRFNFLLESIQRK 1364
KS+A + + + I P KYLG L + + + LL I+
Sbjct: 736 KSQAFLYTN-NRQTESQIMSELPFTIASKRIKYLGIQLTRDVKDLFKENYKPLLNEIKED 794
Query: 1365 LGSWRANMLSLAGRVCLAKSVIAAMPTYTMQV--FNMPRGVTCKINQLIRSFIWSGKGGG 1422
W+ S GR+ + K I Y +P ++ + FIW+ K
Sbjct: 795 TNKWKNIPCSWVGRINIVKMAILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIWNQKRA- 853
Query: 1423 HGWHLVNWEKITQPKDRGGLGVRDTELANTALMGKAIW-CLMHKPNKLWVRVLSHKYLSN 1481
H+ ++Q GG+ + D +L A + K W ++ W R + + +
Sbjct: 854 ---HIAK-STLSQKNKAGGITLPDFKLYYKATVTKTAWYWYQNRDIDQWNRTEPSEIMPH 909
Query: 1482 SSVLQVQAKPQDSQVW 1497
+ KP+ ++ W
Sbjct: 910 IYNYLIFDKPEKNKQW 925
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 116 bits (290), Expect = 7e-25
Identities = 111/408 (27%), Positives = 189/408 (46%), Gaps = 33/408 (8%)
Query: 1018 VRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLLDI 1077
+ + ++R + +S++S +PG DG P ++ + + + +++ G + ++
Sbjct: 344 ITEQDLRASRVSLSS--SPGPDGITPKSAREVPSGIMLRIMNLILWC---GNLPHSIRLA 398
Query: 1078 LVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTM 1137
V IPK +D RPIS+ +V + + +L RL + P Q FLP G
Sbjct: 399 RTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSINW--DPRQRGFLPTDGCA 456
Query: 1138 DNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMS 1197
DNA + V+ H K R +D+ KA+DS+S + + +TL YG P+ ++ + +
Sbjct: 457 DNATIVDLVLRHSHKHF--RSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQN 514
Query: 1198 SVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIR 1257
+ S+ +G F P RG++QGDP+SP LF L M+RL + S +
Sbjct: 515 TYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEIG-------- 566
Query: 1258 LSKNGPPISH-LFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIVSKGV 1316
+K G I++ FADD++LF E + LL T+ F GLK+N K + KG
Sbjct: 567 -AKVGNAITNAAAFADDLVLFAETRMGLQVLLDKTLD-FLSIVGLKLNADKCFTVGIKGQ 624
Query: 1317 SE---VVKDSISVV---APIPFVH--DLGKYLGFPLKG-GRIHRNRFNFLLESIQRKLGS 1367
+ V ++ S + IP + D KYLG GR+ N E I KL
Sbjct: 625 PKQKCTVLEAQSFYVGSSEIPSLKRTDEWKYLGINFTATGRVRCNP----AEDIGPKLQR 680
Query: 1368 WRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFI 1415
L R+ ++V+ + + + ++ GV K ++LIR ++
Sbjct: 681 LTKAPLKPQQRLFALRTVLIPQLYHKLALGSVAIGVLRKTDKLIRYYV 728
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 82.0 bits (201), Expect = 1e-14
Identities = 78/297 (26%), Positives = 136/297 (45%), Gaps = 18/297 (6%)
Query: 1016 QPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNENLL 1075
+P+ ++E++ A+ +APG+DG + H+++ G V
Sbjct: 4 RPIAREEIQCAIKGWKP-SAPGSDGLTVQAITRTRLPRNFVQLHLLR-----GHVPTPWT 57
Query: 1076 DILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRG 1135
+ LIPK + RPI++ + +++ ++L RL + + P Q + G
Sbjct: 58 AMRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYARIDG 115
Query: 1136 TMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLI 1195
T+ N+ L I S+ ++ +D+ KA+D+VS S + L+ G E T N I
Sbjct: 116 TLVNSLLLDTYIS--SRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYI 173
Query: 1196 MSSVTSSQVSI-LWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWK 1254
S++ S +I + G + RG++QGDP+SP+LF ++ L +QS G
Sbjct: 174 TGSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQSTPGIGG-- 231
Query: 1255 PIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAI 1311
+ + P+ L FADD+LL + V LA T+ F G+ +N KS +I
Sbjct: 232 --TIGEEKIPV--LAFADDLLLLEDNDVLLPTTLA-TVANFFRLRGMSLNAKKSVSI 283
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 81.6 bits (200), Expect = 2e-14
Identities = 71/266 (26%), Positives = 114/266 (42%), Gaps = 18/266 (6%)
Query: 1014 LLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNEN 1073
L +P+ DE++ + TA G DG WN + + + + G+
Sbjct: 315 LWRPISNDEIKE--VEACKRTAAGPDGMTT----TAWNSIDECIKSLFNMIMYHGQCPRR 368
Query: 1074 LLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPG 1133
LD VLIPK RP+S+ +VA + ++L NR+ L+ Q +F+
Sbjct: 369 YLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLDTRQRAFIVA 426
Query: 1134 RGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTIN 1193
G +N L +I +G+ +D++KA+DSV + + L P N
Sbjct: 427 DGVAENTSLLSAMIKEARMKI--KGLYIAILDVKKAFDSVEHRSILDALRRKKLPLEMRN 484
Query: 1194 LIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDW 1253
IM +S+ + + +P RG+RQGDP+SP LF M+ +VL + + G
Sbjct: 485 YIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCVMD--AVLRRLPENTG-- 540
Query: 1254 KPIRLSKNGPPISHLFFADDVLLFCE 1279
I L FADD++L E
Sbjct: 541 ----FLMGAEKIGALVFADDLVLLAE 562
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 81.3 bits (199), Expect = 2e-14
Identities = 43/142 (30%), Positives = 73/142 (51%), Gaps = 2/142 (1%)
Query: 1388 AMPTYTMQVFNMPRGVTCKINQLIRSFIWSGKGGGHGWHLVNWEKITQPK-DRGGLGVRD 1446
A+P Y M F + + + K+ + F WS V W+K+ + K D GGLG RD
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 1447 TELANTALMGKAIWCLMHKPNKLWVRVLSHKYLSNSSVLQVQAKPQDSQVWKGLLKARDR 1506
N AL+ K + ++H+P+ L R+L +Y +SS+++ + S W+ ++ R+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 1507 LHTGFVFRLGNG-ETSLWHDDW 1527
L G + +G+G T +W D W
Sbjct: 122 LSRGLLRTIGDGIHTKVWLDRW 143
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 70.9 bits (172), Expect = 3e-11
Identities = 35/65 (53%), Positives = 42/65 (63%)
Query: 1209 NGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHL 1268
NG P RGLRQGDP+SPYLF+LC E LS L + ++G IR+S N P I+HL
Sbjct: 15 NGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRINHL 74
Query: 1269 FFADD 1273
FADD
Sbjct: 75 LFADD 79
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 66.2 bits (160), Expect = 8e-10
Identities = 99/418 (23%), Positives = 170/418 (39%), Gaps = 47/418 (11%)
Query: 1014 LLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQ-EGKVNE 1072
L PV E++ A S P DG P + WN + D ++ F G+V
Sbjct: 155 LWDPVSLIEIKSARASNEKGAGP--DGVTP----RSWNALDDRYKRLLYNIFVFYGRVPS 208
Query: 1073 NLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLP 1132
+ V PK++ RP+S+C+V + K+L R Q+++LP
Sbjct: 209 PIKGSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILARRFVSCY--TYDERQTAYLP 266
Query: 1133 GRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTI 1192
G N + +I +K K +A +DL KA++SV S L + + G P +
Sbjct: 267 IDGVCINVSMLTAIIAE-AKRLRKELHIAI-LDLVKAFNSVYHSALIDAITEAGCPPGVV 324
Query: 1193 NLIMSSVTSSQVSILWNG-CRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKG 1251
+ I + + + G C L + G+ QGDP+S LF L E+ +++L ++G
Sbjct: 325 DYIADMYNNVITEMQFEGKCELASIL--AGVYQGDPLSGPLFTLAYEK---ALRALNNEG 379
Query: 1252 --DWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSK 1309
D +R++ + ++DD LL + + L + GL+IN KSK
Sbjct: 380 RFDIADVRVNASA-------YSDDGLLLAMTVIGLQHNLDKFGETLA-KIGLRINSRKSK 431
Query: 1310 --AIVSKGVSEVVKD--------SISVVAPIPFVHDLGKYLG--FPLKGGRIHRNRFNFL 1357
++V G + +K S + P+ + DL KYLG + G + +
Sbjct: 432 TVSLVPSGREKKMKIVSNRRLLLKASELKPLT-ISDLWKYLGVVYTTSGPEVAK------ 484
Query: 1358 LESIQRKLGSWRANMLSLAGRVCLAKSVIAAMPTYTMQVFNMPRGVTCKINQLIRSFI 1415
S+ L L R+ L K+ + + + CK++ LIR ++
Sbjct: 485 -VSMDDDLSKLTKGPLKPQQRIHLLKTFVIPKHLNRLVLSRTTATGLCKMDLLIRKYV 541
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 65.5 bits (158), Expect = 1e-09
Identities = 58/216 (26%), Positives = 97/216 (44%), Gaps = 20/216 (9%)
Query: 1135 GTMDNAFLAQEVIHHMSKSTAKRGMV--AFKIDLEKAYDSVSWSFLQETLELYGFPEVTI 1192
GT+ N + + H K +G +D+ KA+D+VS + + +G +
Sbjct: 1 GTLANIIM----LEHYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQ 56
Query: 1193 NLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSLVDKGD 1252
+ IMS++T + +I+ G G++QGDP+SP LF + ++ L + D
Sbjct: 57 DFIMSTITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRL------ND 110
Query: 1253 WKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLSKSKAIV 1312
+P I+ L FADD+LL + + N LA+T F + G+ +N K +I
Sbjct: 111 EQPGASMTPACKIASLAFADDLLLLEDRDIDVPNSLATTCAYF-RTRGMTLNPEKCASIS 169
Query: 1313 SKGVS--EVVKDSISVVAPIPFVHDLG-----KYLG 1341
+ VS V + S ++ LG KYLG
Sbjct: 170 AATVSGRSVPRSKPSFTIDGRYIKPLGGINTFKYLG 205
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 64.7 bits (156), Expect = 2e-09
Identities = 51/216 (23%), Positives = 94/216 (42%), Gaps = 20/216 (9%)
Query: 1063 EAFQEGKVNENLLDILVVLIPKVDQPSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKL 1122
++F + + +++ IPK PS+ + RPISL + +++ +++ +R+R L
Sbjct: 654 QSFSDSAIPNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHL 713
Query: 1123 IGPMQSSFLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETL 1182
+ P Q FL R + + + H + K+ ++ F D KA+D VS L + L
Sbjct: 714 LSPHQHGFLNFRSCPSSLVRSISLYHSILKNEKSLDILFF--DFAKAFDKVSHPILLKKL 771
Query: 1183 ELYGFPEVTINLIMSSVTSSQVSILWNG-CRLPAFKPGRGLRQGDPMSPYLFVLCMERLS 1241
L+G ++T + + S+ N A+ G+ QG P LF+L + L
Sbjct: 772 ALFGLDKLTCSWFKEFLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGPLLFILFINDLL 831
Query: 1242 VLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLF 1277
+ ++ P I FADD+ +F
Sbjct: 832 IDLE-----------------PNIHVSCFADDIKIF 850
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 47.8 bits (112), Expect = 3e-04
Identities = 52/223 (23%), Positives = 92/223 (40%), Gaps = 10/223 (4%)
Query: 1021 DEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVNEN-LLDILV 1079
DEV ++ +PG DG + W + + ++ + K+ E + + LV
Sbjct: 408 DEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIASLV 467
Query: 1080 VLIPKVDQ-PSTVRDLRPISLCNVAFKVITKVLVNRLRPFLRKL-IGPMQSSFLPGRGTM 1137
+L+ +D+ S RPI L + KV+ ++V RL L + + P Q +F G+ T
Sbjct: 468 ILLKLLDRIRSDPGSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKSTE 527
Query: 1138 DNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMS 1197
D A + + + + ++ ID + A+D++ W L L+L L MS
Sbjct: 528 D----AWRCVQRHVECSEMKYVIGLNIDFQGAFDNLGW--LSMLLKLDEAQSNEFGLWMS 581
Query: 1198 SVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERL 1240
+V + + RG QG P ++ L M L
Sbjct: 582 YFGGRKVYYV-GKTGIVRKDVTRGCPQGSKSGPAMWKLVMNEL 623
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 46.2 bits (108), Expect = 9e-04
Identities = 82/361 (22%), Positives = 139/361 (37%), Gaps = 43/361 (11%)
Query: 1001 DLFPSLSVEAKSLLLQPVRKD----EVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDS 1056
+ FP EA + + + V EV + + S +PG DG K W + +
Sbjct: 413 NFFPVAESEAPTAIAEEVPPALEVFEVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEH 472
Query: 1057 LWHMVKEAFQEGKVNENLLDILVVLI---PKVD--QPSTVRDLRPISLCNVAFKVITKVL 1111
L + + G VV + P D +PS+ R I L V KV+ ++
Sbjct: 473 LASLFSRCIRLGYFPAEWKCPRVVSLLKGPDKDKCEPSSYRG---ICLLPVFGKVLEAIM 529
Query: 1112 VNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYD 1171
VNR+R L + Q F GR D A + ++A + ++ +D + A+D
Sbjct: 530 VNRVREVLPEGC-RWQFGFRQGRCVED----AWRHVKSSVGASAAQYVLGTFVDFKGAFD 584
Query: 1172 SVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPY 1231
+V WS L G E+ + S S + +++ + RG QG P+
Sbjct: 585 NVEWSAALSRLADLGCREMG---LWQSFFSGRRAVIRSSSGTVEVPVTRGCPQGSISGPF 641
Query: 1232 LFVLCMERLSVLIQSLVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQV-----N 1286
++ + M+ VL+Q L P +ADD+LL E + V
Sbjct: 642 IWDILMD---VLLQRL--------------QPYCQLSAYADDLLLLVEGNSRAVLEEKGA 684
Query: 1287 LLASTMQLFCDSSGLKINLSKSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKG 1346
L S ++ + G ++ SK+ ++ KG +P+V +YLG +
Sbjct: 685 QLMSIVETWGAEVGDCLSTSKTVIMLLKGALRRAPTVRFAGRNLPYVRSC-RYLGITVSE 743
Query: 1347 G 1347
G
Sbjct: 744 G 744
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 42.7 bits (99), Expect = 0.010
Identities = 128/645 (19%), Positives = 233/645 (35%), Gaps = 76/645 (11%)
Query: 800 VDVLNRVHSDHCPILLQCGSPQTLAHNRPFR--FLAAWADHPDFAAV------------- 844
V L+ + SDH P+L + N+P R LA AD F A
Sbjct: 204 VQTLHELSSDHTPLLADL---HAMPINKPPRSCLLARGADIERFKAYLTQHIDLSVGIQG 260
Query: 845 ---VDNAW--------RSGEECITSKLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQ 893
+DNA R+ S + V + + + ++ R KR V +
Sbjct: 261 TDDIDNAIDSFMDILKRAAIRSAPSHQQNVESSRQLQLPPIVASLIRLKRKV-------R 313
Query: 894 RELDRRVTSDMVLFEAELQREYRNILRQEELL*YQKARENRVHLGDRNTAYFHTQTLIRR 953
RE R + + + L +L + + EN G N + + +
Sbjct: 314 REYARTGDARIQQIHSRLANRLHKVLNRRKQSQIDNLLENLDTDGSTNFSLWRITKRYKT 373
Query: 954 RKNRVHRLKLEDGTWCGDEEVLKRVVHGFFVNLFTGITPLASPRVTHDLFPSLSVEAK-- 1011
+ ++ G WC V F +L LA +H L + S++
Sbjct: 374 QATPNSAIRNPAGGWCRTSREKTEV---FANHLEQRFKALAFAPESHSLMVAESLQTPFQ 430
Query: 1012 -SLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKV 1070
+L PV +EV+ + + APG D + ++ L + + G
Sbjct: 431 MALPADPVTLEEVKELVSKLKPKKAPGEDLLDNRTIRLLPDQALLYLVLIFNSILRVGYF 490
Query: 1071 NENLLDILVVLIPKV-DQPSTVRDLRPISLCNVAFKVITKVLVNRL--RPFLRKLIGPMQ 1127
+ +++I K QP V RP SL K++ ++++NR+ + + I Q
Sbjct: 491 PKARPTASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRILTSEEVTRAIPKFQ 550
Query: 1128 SSFLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGF 1187
F GT + L + V + K + +D+++A+D V L +
Sbjct: 551 FGFRLQHGTPEQ--LHRVVNFALEALEKKEYAGSCFLDIQQAFDRVWHPGLLYKAKSLLS 608
Query: 1188 PEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQSL 1247
P++ LI S + S+ +GCR G+ QG + P L+ +
Sbjct: 609 PQL-FQLIKSFWEGRKFSVTADGCRSSVKFIEAGVPQGSVLGPTLYSI----------FT 657
Query: 1248 VDKGDWKPIRLSKNGPPISHLFFADDVLLFCEAS--VAQVNLLASTMQLFCDSS---GLK 1302
D + + G + +ADD+ + +++ V + L + F + + +
Sbjct: 658 ADMPNQNAVTGLAEG-EVLIATYADDIAVLTKSTCIVEATDALQEYLDAFQEWAVKWNVS 716
Query: 1303 INLSKSKAIVSKGVSEVVKDSISVV---APIPFVHDLGKYLGFPLKGGRIHRNRFNFLLE 1359
IN K + + ++D V + + H+ KYLG L R L
Sbjct: 717 INAGKCANVT---FTNAIRDCPGVTINGSLLSHTHEY-KYLGVILDRSLTFRRHITSLQH 772
Query: 1360 SIQRKLG--SW---RANMLSLAGRVCLAKSVIAAMPTYTMQVFNM 1399
S + ++ +W N LSL +V + K ++A Y +QV+ +
Sbjct: 773 SFRTRITKMNWLLAARNKLSLDNKVKIYKCIVAPGLFYAIQVYGI 817
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 42.4 bits (98), Expect = 0.012
Identities = 80/398 (20%), Positives = 156/398 (39%), Gaps = 22/398 (5%)
Query: 1012 SLLLQPVRKDEVRHALMSMNSYTAPGADGFQPFFYKKYWNRVGDSLWHMVKEAFQEGKVN 1071
SL L V +EV++ + + APG D + ++ L + G
Sbjct: 432 SLPLSAVTLEEVKNLIAKLPLKKAPGEDLLDNRTIRLLPDQALQFLALIFNSVLDVGYFP 491
Query: 1072 ENLLDILVVLIPKVDQ-PSTVRDLRPISLCNVAFKVITKVLVNRLRPF--LRKLIGPMQS 1128
+ +++I K + P+ V RP SL K++ ++++NRL + K I Q
Sbjct: 492 KAWKSASIIMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQF 551
Query: 1129 SFLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSW--SFLQETLELYG 1186
F GT + L + V + K V +D+++A+D V W L + L
Sbjct: 552 GFRLQHGTPEQ--LHRVVNFALEAMENKEYAVGAFLDIQQAFDRV-WHPGLLYKAKRL-- 606
Query: 1187 FPEVTINLIMSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLFVLCMERLSVLIQS 1246
FP ++ S + + +G + G+ QG + P L+ + + +
Sbjct: 607 FPPQLYLVVKSFLEERTFHVSVDGYKSSIKPIAAGVPQGSVLGPTLYSVFASDMPT--HT 664
Query: 1247 LVDKGDWKPIRLSKNGPPISHLFFADDVLLFCEASVAQVNLLASTMQLFCDSSGLKINLS 1306
V + D + + ++ + L + +L A+ + + Q + ++ ++IN
Sbjct: 665 PVTEVDEEDVLIATYADDTAVLTKSKSIL----AATSGLQEYLDAFQQWAENWNVRINAE 720
Query: 1307 KSKAIVSKGVSEVVKDSISVVAPIPFVHDLGKYLGFPLKGGRIHRNRFNFLLESIQRKLG 1366
K + + +S+ + H KYLG L + ++ + K+
Sbjct: 721 KCANVTFANRTGSCP-GVSLNGRLIRHHQAYKYLGITLDRKLTFSRHITNIQQAFRTKVA 779
Query: 1367 --SWRA---NMLSLAGRVCLAKSVIAAMPTYTMQVFNM 1399
SW N LSL +V + KS++A Y +QV+ +
Sbjct: 780 RMSWLIAPRNKLSLGCKVNIYKSILAPCLFYGLQVYGI 817
>RTL_CHLRE (P11660) Reverse transcriptase-like protein
Length = 368
Score = 38.5 bits (88), Expect = 0.18
Identities = 40/148 (27%), Positives = 57/148 (38%), Gaps = 14/148 (9%)
Query: 1094 LRPISLCNVAFKVITKVLVNRLRPFLRKLIGPMQSSFLPGRGTMDNAFLAQEVIHHMSKS 1153
+RPI + + ++ V +NRL F PG GT + M
Sbjct: 117 VRPIIVAHKGLRICMAV-INRLLQSCCVSWSNQTFGFRPGFGTHHAVLHLAQKAQQMLNK 175
Query: 1154 TAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPEVTINLIMSSVTSSQVSILWNGCRL 1213
+ +V F DL+ AY+SV L +TL+L P LI +W L
Sbjct: 176 QQQAVLVTF--DLQAAYNSVDIKHLMQTLQLQYLPNDMKKLIW----------MWQHLPL 223
Query: 1214 PAFKPG-RGLRQGDPMSPYLFVLCMERL 1240
G GL QG SP LF +++L
Sbjct: 224 AQLNAGINGLAQGYAYSPTLFAWYVDQL 251
>RT16_MYXXA (P23072) RNA-directed DNA polymerase from retron Mx162-RT
(EC 2.7.7.49) (Reverse transcriptase) (Mx162-RT)
Length = 485
Score = 37.7 bits (86), Expect = 0.31
Identities = 36/123 (29%), Positives = 55/123 (44%), Gaps = 17/123 (13%)
Query: 1130 FLPGRGTMDNAFLAQEVIHHMSKSTAKRGMVAFKIDLEKAYDSVSWSFLQETLELYGFPE 1189
F+ GR + NA Q V K+DL+ + SV+W ++ L G E
Sbjct: 227 FVAGRSILTNALAHQGAD------------VVVKVDLKDFFPSVTWRRVKGLLRKGGLRE 274
Query: 1190 VTINLI-MSSVTSSQVSILWNGCRLPAFKPGRGLRQGDPMSPYLF-VLCM---ERLSVLI 1244
T L+ + S + + ++ + G L K R L QG P SP + LC+ +RLS L
Sbjct: 275 GTSTLLSLLSTEAPREAVQFRGKLLHVAKGPRALPQGAPTSPGITNALCLKLDKRLSALA 334
Query: 1245 QSL 1247
+ L
Sbjct: 335 KRL 337
>OFD1_HUMAN (O75665) Oral-facial-digital syndrome 1 protein (Protein
71-7A)
Length = 1012
Score = 36.2 bits (82), Expect = 0.89
Identities = 27/114 (23%), Positives = 53/114 (45%), Gaps = 3/114 (2%)
Query: 852 GEECITSKLERVREASTVFNKEVFGNIFRRKRHVEARLRGVQRELDRRVTSDMVLFEAEL 911
G E + E + +E +I R E ++ + DR++ ++++ ++ EL
Sbjct: 298 GREAELKQRVEAFELNQKLQEEKHKSITEALRRQEQNIKSFEETYDRKLKNELLKYQLEL 357
Query: 912 QREYRNILRQEELL*YQ-KARENRVHLGDRNTAYFHTQTLIRRRKNRVHRLKLE 964
+ +Y I+R L+ + K +E VHL + A + + + NRV L+LE
Sbjct: 358 KDDY--IIRTNRLIEDERKNKEKAVHLQEELIAINSKKEELNQSVNRVKELELE 409
>GNT2_RAT (Q09326) Alpha-1,6-mannosyl-glycoprotein
2-beta-N-acetylglucosaminyltransferase (EC 2.4.1.143)
(N-glycosyl-oligosaccharide-glycoprotein
N-acetylglucosaminyltransferase II)
(Beta-1,2-N-acetylglucosaminyltransferase II) (GNT-II)
(GlcNAc-T II)
Length = 442
Score = 35.4 bits (80), Expect = 1.5
Identities = 28/98 (28%), Positives = 44/98 (44%), Gaps = 5/98 (5%)
Query: 1583 VPNRQDVWTWET--NSMGIYTVRGAYRWLQDQQSHLPVVDDWNWIWKLK-VPEKIRTFVW 1639
+ ++ DV TW++ ++MG+ R AY+ L + DD+NW W L+ + VW
Sbjct: 298 IADKVDVKTWKSTEHNMGLALTRDAYQKLIECTDTFCTYDDYNWDWTLQYLTLACLPKVW 357
Query: 1640 LTLQNSLQVNLHRFRCKMAASPSC--SRCSAPEEDILH 1675
L H C M +C S SA E +L+
Sbjct: 358 KVLVPQAPRIFHAGDCGMHHKKTCRPSTQSAQIESLLN 395
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.332 0.143 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 221,219,800
Number of Sequences: 164201
Number of extensions: 9301677
Number of successful extensions: 20962
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 20885
Number of HSP's gapped (non-prelim): 64
length of query: 1898
length of database: 59,974,054
effective HSP length: 125
effective length of query: 1773
effective length of database: 39,448,929
effective search space: 69942951117
effective search space used: 69942951117
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0114.7


