

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0113.5
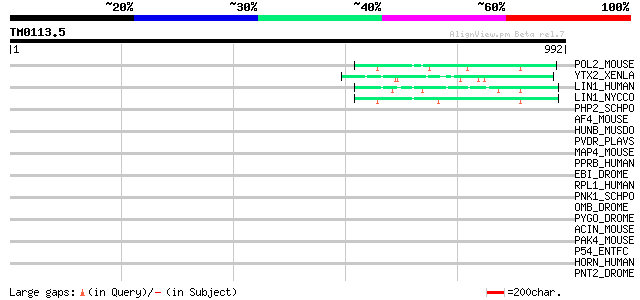
(992 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 63 3e-09
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 60 3e-08
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 54 2e-06
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 50 3e-05
PHP2_SCHPO (P24488) Transcriptional activator php2 40 0.040
AF4_MOUSE (O88573) AF-4 protein (Proto-oncogene AF4) 38 0.12
HUNB_MUSDO (Q01778) Hunchback protein 37 0.20
PVDR_PLAVS (P22290) Duffy receptor precursor (Erythrocyte bindin... 37 0.34
MAP4_MOUSE (P27546) Microtubule-associated protein 4 (MAP 4) 36 0.45
PPRB_HUMAN (Q15648) Peroxisome proliferator-activated receptor b... 36 0.58
EBI_DROME (Q95RJ9) F-box-like/WD-repeat protein ebi 36 0.58
RPL1_HUMAN (Q8IWN7) Retinitis pigmentosa 1-like 1 protein 35 0.99
PNK1_SCHPO (O13911) Bifunctional polynucleotide phosphatase/kina... 35 0.99
OMB_DROME (Q24432) Optomotor-blind protein (Lethal(1)optomotor-b... 35 0.99
PYGO_DROME (Q9V9W8) Pygopus protein (Gammy legs protein) 35 1.3
ACIN_MOUSE (Q9JIX8) Apoptotic chromatin condensation inducer in ... 35 1.3
PAK4_MOUSE (Q8BTW9) Serine/threonine-protein kinase PAK 4 (EC 2.... 34 1.7
P54_ENTFC (P13692) P54 protein precursor 34 1.7
HORN_HUMAN (Q86YZ3) Hornerin 34 1.7
PNT2_DROME (P51023) ETS-like protein pointed, isoform P2 (D-ETS-2) 34 2.2
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 63.2 bits (152), Expect = 3e-09
Identities = 74/383 (19%), Positives = 152/383 (39%), Gaps = 25/383 (6%)
Query: 617 SPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASD-----KMGGGPPNLLSMAKFR 671
+PNA F+ L + + + + VGDFNT L + D K+ L + K
Sbjct: 142 APNARAATFIRDTLVKLKAYIAPHTIIVGDFNTPLSSKDRSWKQKLNRDTVKLTEVMKQM 201
Query: 672 DCLDACSLSDLGFKGPPFTWKGRGVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPI 731
D D KG F G +ID +G+ ++ + ++ P + DH +
Sbjct: 202 DLTDIYRTFYPKTKGYTFFSAPHGTFSKIDHIIGHKTGLNRYKNIEIV--PCILSDHHGL 259
Query: 732 LIQLGPEHMDSSQRPFRF------LAAWLTHEDFPRLVKSSWDSHHDSWIPASEAFCEEA 785
+ ++++ + F + L L E + +K + + + +
Sbjct: 260 RLIFN-NNINNGKPTFTWKLNNTLLNDTLVKEGIKKEIKDFLEFNENEATTYPNLWDTMK 318
Query: 786 ASWNRDVFGEIGRTKRRLMRRLEGINNHLRM-----SHSPYLEKLQK--RLWKEYQTVLI 838
A + K+R + HL+ ++SP + Q+ +L E V
Sbjct: 319 AFLRGKLIALSASKKKRETAHTSSLTTHLKALEKKEANSPKRSRRQEIIKLRGEINQVET 378
Query: 839 REELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAV 898
R + ++R + ++ T HR + I + N+ G++ TD +++++
Sbjct: 379 RRTIQRINQTRSWFFEKINKIDKPLARLTKGHRDKILINKIRNEKGDITTDPEEIQNTIR 438
Query: 899 TSFQNLYTSPGPG----EPYHVRNAFPSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPG 954
+ ++ LY++ + + R P L ++ +D + P+ EI + S+ K+PG
Sbjct: 439 SFYKRLYSTKLENLDEMDKFLDRYQVPKLNQDQVDHLNSPISPKEIEAVINSLPTKKSPG 498
Query: 955 PDDLNPLFFQSQWQVIGPSVVKL 977
PD + F+Q+ + + P + KL
Sbjct: 499 PDGFSAEFYQTFKEDLIPILHKL 521
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 60.1 bits (144), Expect = 3e-08
Identities = 94/412 (22%), Positives = 166/412 (39%), Gaps = 49/412 (11%)
Query: 594 QFVHLRIHPKNNVPNFLVTFV--YGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYL 651
+ +HLR+ N + + G A L ++ I S + + GDFN L
Sbjct: 91 RLLHLRVRESGRTYNLMNVYAPTTGPERARFFESLSAYMETIDSD--EALIIGGDFNYTL 148
Query: 652 QASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPP----FTW----KGRGVKERIDWA 703
A D+ + S + R+ + SL D+ + P FT+ G + RID
Sbjct: 149 DARDRNVPKKRDS-SESVLRELIAHFSLVDVWREQNPETVAFTYVRVRDGHVSQSRIDRI 207
Query: 704 LGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLV 763
++ +S +T+ P ++ + + + P ++ + F + L E F + V
Sbjct: 208 YISSHLMSRAQSSTIRLAPFSDHNCVSLRMSIAPSLPKAAY--WHFNNSLLEDEGFAKSV 265
Query: 764 KSSWDSHHDSWIPASEAFCEEAASWNRDVFGEIGRTKRRLM-----RRLEGINN-HLRMS 817
+ +W W AF +E A+ N+ + ++G+ +L+ + + G N +
Sbjct: 266 RDTWRG----W----RAFQDEFATLNQ--WWDVGKVHLKLLCQEYTKSVSGQRNAEIEAL 315
Query: 818 HSPYLEKLQKRLWKEYQTV----LIREELLWRQ----------KSRVNWLNHGDRNTHFF 863
+ L+ Q+ E Q + L R+E L +SR+ L DR + FF
Sbjct: 316 NGEVLDLEQRLSGSEDQALQCEYLERKEALRNMEQRQARGAFVRSRMQLLCDMDRGSRFF 375
Query: 864 HTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNAFPSL 923
+ R +I L + G + D + +R A + +QNL+ SP P P + L
Sbjct: 376 YALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNLF-SPDPISPDACEELWDGL 434
Query: 924 P---RETLDEIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGP 972
P + + P+ E+ A+ M K+PG D L FFQ W +GP
Sbjct: 435 PVVSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFFQFFWDTLGP 486
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 54.3 bits (129), Expect = 2e-06
Identities = 74/401 (18%), Positives = 161/401 (39%), Gaps = 53/401 (13%)
Query: 617 SPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDA 676
+PN F+ Q L ++ + + +GDFNT L D+ N + + L
Sbjct: 115 APNTGAPRFIKQVLSDLQRDLDSHTIIMGDFNTPLSTLDRSTRQKINK-DIQELNSALHQ 173
Query: 677 CSLSDLG------------FKGPPFTWKGRGVKERIDWALGNARWISSFPEATVLHLPKL 724
L D+ F P T+ + D LG+ +S ++
Sbjct: 174 ADLIDIYRTLHPKSTEYTFFSAPHHTYS------KTDHILGSKTLLSKCKRTEII--TNC 225
Query: 725 KYDHKPILIQLG-----PEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDSWIPASE 779
DH I ++L H + + L + H + +K ++++ +
Sbjct: 226 LSDHSAIKLELRIKKLTQNHSTTWKLNNLLLNDYWVHNEMKAEIKKFFETNENKDTTYQN 285
Query: 780 AFCEEAASWNRDVFGEIGRTKRRLMR-RLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLI 838
+ + A + R F + KR+ R +++ + + L+ E+ + + + + I
Sbjct: 286 LW-DTAKAVCRGKFIALNAHKRKQERSKIDTLISQLKELEKQ--EQTNSKASRRQEIIKI 342
Query: 839 REEL--LWRQKS--RVNWLNHGDRNTHFFHTTTMVHR----------KRNKIEALNNDVG 884
R EL + QK+ ++N + + FF + R ++N+I+ + ND G
Sbjct: 343 RAELKEIETQKTLQKIN-----ESRSWFFEKINKIDRPLARLIKKKREKNQIDTIKNDRG 397
Query: 885 ELITDQDQLRSMAVTSFQNLYTSPGPG----EPYHVRNAFPSLPRETLDEIMGPLEAAEI 940
++ TD ++++ +++LY + + + P L +E ++ + P+ ++EI
Sbjct: 398 DITTDPTEIQTTIREYYKHLYANKLENLEEMDKFLDTYTLPRLNQEEVESLNRPITSSEI 457
Query: 941 RVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVVKLAMTV 981
+ S+ K+PGP+ F+Q + + P ++KL ++
Sbjct: 458 EAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQSI 498
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 50.1 bits (118), Expect = 3e-05
Identities = 70/386 (18%), Positives = 149/386 (38%), Gaps = 23/386 (5%)
Query: 617 SPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASD-----KMGGGPPNLLSMAKFR 671
+PN + F+ + L ++++ ++ + VGDFNT L D K+ +L S +
Sbjct: 115 APNHNAPQFIRETLTDMSNLISSTSIVVGDFNTPLAVLDRSSKKKLSKEILDLNSTIQHL 174
Query: 672 DCLDACSLSDLGFKGPPFTWKGRGVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPI 731
D D F G +ID LG+ +S F + ++ P + DH I
Sbjct: 175 DLTDIYRTFHPNKTEYTFFSSAHGTYSKIDHILGHKSNLSKFKKIEII--PCIFSDHHGI 232
Query: 732 LIQLGPE---HMDSSQRPFRFLA---AWLTHEDFPRLVK-----SSWDSHHDS-WIPASE 779
++L H + L W+ E + K ++ D+++ + W A
Sbjct: 233 KVELNNNRNLHTHTKTWKLNNLMLKDTWVIDEIKKEITKFLEQNNNQDTNYQNLWDTAKA 292
Query: 780 AFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIR 839
+ + + LM L+ + + P K ++ E + +
Sbjct: 293 VLRGKFIALQAFLKKTEREEVNNLMGHLKQLEKEEHSNPKPSRRKEITKIRAELNEIENK 352
Query: 840 EELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVT 899
+ KS+ + ++ T R ++ I ++ N E+ TD +++ +
Sbjct: 353 RIIQQINKSKSWFFEKINKIDKPLANLTRKKRVKSLISSIRNGNDEITTDPSEIQKILNE 412
Query: 900 SFQNLYTSPGPG----EPYHVRNAFPSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGP 955
++ LY+ + Y P L ++ ++ + P+ ++EI + ++ K+PGP
Sbjct: 413 YYKKLYSHKYENLKEIDQYLEACHLPRLSQKEVEMLNRPISSSEIASTIQNLPKKKSPGP 472
Query: 956 DDLNPLFFQSQWQVIGPSVVKLAMTV 981
D F+Q+ + + P ++ L +
Sbjct: 473 DGFTSEFYQTFKEELVPILLNLFQNI 498
>PHP2_SCHPO (P24488) Transcriptional activator php2
Length = 334
Score = 39.7 bits (91), Expect = 0.040
Identities = 41/186 (22%), Positives = 69/186 (37%), Gaps = 28/186 (15%)
Query: 271 QAAANQTKHSGGKGAETRFSILEQMDVDAIHGSSGIGASSSGILA*VQIPEQNNLNQNTP 330
Q AA + G G + + V A SS + +S G + N+ ++
Sbjct: 76 QEAAEAAANGGSTGDDVNATNANDATVPATV-SSEVTHTSEGYA------DSNDSRPSSI 128
Query: 331 TNQESQPQHMVDSAGSGISPTKGKVVNTNDEPNHEGISSVPNDETMISGPTKITSAPTGP 390
+N P ++SA + +SP N PN G + + M GPT T++ +GP
Sbjct: 129 SNSSESPAP-INSATASMSPANNTSGNNITSPNVRGELDMSGNIAMSGGPTN-TASTSGP 186
Query: 391 QPHDGGTGTKVNPNSQGAIPQQSR-------------------ADVAHPSHFKSGQHDKL 431
PHD + + N+ + S+ + AHP F G +D
Sbjct: 187 VPHDMTVLPQTDSNTSNLMSSGSQLGSFATASTNGNNSTTTTTSSAAHPGSFHKGTNDYS 246
Query: 432 GPTSGS 437
+G+
Sbjct: 247 STLAGN 252
>AF4_MOUSE (O88573) AF-4 protein (Proto-oncogene AF4)
Length = 1217
Score = 38.1 bits (87), Expect = 0.12
Identities = 47/186 (25%), Positives = 73/186 (38%), Gaps = 30/186 (16%)
Query: 326 NQNTPTNQESQPQHMVDSAGSGISP-----TKGKVVNTNDEPNHEGISSVPNDETM-ISG 379
N T NQ S P +D GS SP +KG ++D+ + + +P + + G
Sbjct: 504 NWLTKVNQPSVP---LDGRGSTESPQWRQESKGVAEGSSDQQHPDSKDPLPKSSSKTLRG 560
Query: 380 PTK--------ITSAPTGPQPHDGGTGTKVNPNSQGAIPQQSRADVAHPSHFKSGQHDKL 431
P++ + + P PH G NP + P + R A S S ++
Sbjct: 561 PSEGPSLGRGAVRNPPLNRNPHLGKPWAANNPGN----PPRLRPGRAQAS---SQAESEV 613
Query: 432 GP----TSGSSSLKKTKMKARGHQRVEGSSEPRRSPNASSNAPGDMGGRTFRERDRSLSP 487
GP + +S + K+K +G R GS EP+ P + P R
Sbjct: 614 GPLPYRSKEQTSKDRPKVKTKGRPRAVGSREPK--PEVPAPTPQAAVPRPKPPVPTPSEK 671
Query: 488 RKHRSS 493
RKH+SS
Sbjct: 672 RKHKSS 677
>HUNB_MUSDO (Q01778) Hunchback protein
Length = 817
Score = 37.4 bits (85), Expect = 0.20
Identities = 35/165 (21%), Positives = 67/165 (40%), Gaps = 12/165 (7%)
Query: 324 NLNQNTPTNQESQPQHMVDSAGSGISPTKGKVVNTNDEPNHEGISSVPNDETMISGPTKI 383
+L+Q TPT +E Q + + + + +++ +G + D+ +G +I
Sbjct: 605 DLSQGTPTKEEQQTPELAMNLKLSEEHGETPLFSSSAAARRKG-RVLKLDQEKTAGHLQI 663
Query: 384 TSAPTGPQPH-------DGGTGTKVNPNSQGAIPQQSRADVAHPSHFKSG-QHDKLGPTS 435
SAPT PQ H T + ++P+ + + + ++G H PT+
Sbjct: 664 ASAPTSPQHHLHHNNEMPPTTSSPIHPSQVNGVAAGAADHSSADESMETGHHHHHHNPTT 723
Query: 436 GSSSLKKTKMKARGHQRVEGSSEPRRSPNASSNAPGDMGGRTFRE 480
++S T A SS S N++S++ G+ T E
Sbjct: 724 ANTSASST---ASSSGNSSNSSSTSTSSNSNSSSAGNSPNTTMYE 765
>PVDR_PLAVS (P22290) Duffy receptor precursor (Erythrocyte binding
protein)
Length = 1070
Score = 36.6 bits (83), Expect = 0.34
Identities = 49/230 (21%), Positives = 82/230 (35%), Gaps = 10/230 (4%)
Query: 261 DSQENCQNKHQAAANQTKHSGGKGAETRFSILEQMDVDAIHGSSGIGASSSGILA*VQIP 320
++QE N AA +Q +S S E++ D+ HG+ G SS V
Sbjct: 515 NTQEVVTNVDNAAKSQATNSNPISQPVDSSKAEKVPGDSTHGNVNSGQDSSTTGKAVTGD 574
Query: 321 EQNNLNQNTPTNQESQPQHMVDS-AGSGISPTKGKVVNTNDEPNHEGISSVPNDETMISG 379
QN TP + Q + +S + + P K ++D + GI+ + S
Sbjct: 575 GQN--GNQTPAESDVQRSDIAESVSAKNVDPQKSVSKRSDDTASVTGIAEAGKENLGASN 632
Query: 380 PTKITSAPTGPQPHDGGTGTKVNPNSQGAIPQQSRADVAHPSHFKSGQHDKLGPTSG--- 436
S P D + P G P + + S S D GP
Sbjct: 633 SRPSESTVEANSPGDDTVNSASIPVVSGENPLVTPYNGLRHSKDNS---DSDGPAESMAN 689
Query: 437 -SSSLKKTKMKARGHQRVEGSSEPRRSPNASSNAPGDMGGRTFRERDRSL 485
S+ K K + + + + + S + +S+A GD RE ++ +
Sbjct: 690 PDSNSKGETGKGQDNDMAKATKDSSNSSDGTSSATGDTTDAVDREINKGV 739
>MAP4_MOUSE (P27546) Microtubule-associated protein 4 (MAP 4)
Length = 1125
Score = 36.2 bits (82), Expect = 0.45
Identities = 41/181 (22%), Positives = 67/181 (36%), Gaps = 35/181 (19%)
Query: 327 QNTPT-NQESQPQHMVDSAGSGISP------------TKGKVVNTNDEPNHEGISSVPND 373
+ TPT ++ + P +V + S SP T+GK + + +
Sbjct: 775 KTTPTVSKATSPSTLVSTGPSSRSPATTLPKRPTSIKTEGKPADVKRMTAKSASADLSRS 834
Query: 374 ETMISGPTKITSAPTGPQPHDGGTGTKVNP-----NSQGAIPQQSRADVAHPSHFKSGQH 428
+T + K + PTG P G T T+V P S GA+ + PS + +
Sbjct: 835 KTTSASSVKRNTTPTGAAPPAGMTSTRVKPMSAPSRSSGALSVDKKPTSTKPSS-SAPRV 893
Query: 429 DKLGPTSGSSSLKKTKMK-----------ARGHQRVEGSSEP-----RRSPNASSNAPGD 472
+L T + LK + K G +VE +E + PNA + A G
Sbjct: 894 SRLATTVSAPDLKSVRSKVGSTENIKHQPGGGRAKVEKKTEAATTAGKPEPNAVTKAAGS 953
Query: 473 M 473
+
Sbjct: 954 I 954
>PPRB_HUMAN (Q15648) Peroxisome proliferator-activated receptor
binding protein (PBP) (PPAR binding protein) (Thyroid
hormone receptor-associated protein complex 220 kDa
component) (Trap220) (Thyroid receptor interacting
protein 2) (TRIP2) (p53 regulatory
Length = 1581
Score = 35.8 bits (81), Expect = 0.58
Identities = 49/195 (25%), Positives = 77/195 (39%), Gaps = 33/195 (16%)
Query: 308 ASSSGILA*VQIPEQNNLNQNTPTNQES--QPQHMVDSAGSGISPT-------KGKVVNT 358
+SSSG+ + + +L+Q TP + S S+GS +S + KGK +
Sbjct: 1238 SSSSGMKSSSGLGSSGSLSQKTPPSSNSCTASSSSFSSSGSSMSSSQNQHGSSKGKSPSR 1297
Query: 359 NDEPNHEGISSVPNDETMISGPTKITSAPTGPQPHDGGTGTKVNPNSQGAIPQQSRADVA 418
N +P+ + +TS P G P DG G N +S P S+ +++
Sbjct: 1298 NKKPSLTAVIDKLKHGV-------VTSGPGGEDPLDGQMGVSTNSSSH---PMSSKHNMS 1347
Query: 419 ----HPSHFKSGQHDKLGPTSGSS--SLKKT----KMKARGHQRVEGSSEPRRSPNASSN 468
KS + TSGSS S KKT + + G ++ S SP+ +
Sbjct: 1348 GGEFQGKREKSDKDKSKVSTSGSSVDSSKKTSESKNVGSTGVAKIIISKHDGGSPSIKAK 1407
Query: 469 A----PGDMGGRTFR 479
PG+ G R
Sbjct: 1408 VTLQKPGESSGEGLR 1422
>EBI_DROME (Q95RJ9) F-box-like/WD-repeat protein ebi
Length = 700
Score = 35.8 bits (81), Expect = 0.58
Identities = 35/152 (23%), Positives = 55/152 (36%), Gaps = 12/152 (7%)
Query: 303 SSGIGASSSGILA*VQIPEQNNLNQNTPTNQESQPQHMVDSAGSGISPTKGKVVNTNDEP 362
S+ G+SSSG ++ +T TN S V+ SG++P G V +N +
Sbjct: 213 SNEAGSSSSGNAGNANATSTDDAASSTSTNGNSSTSSSVEQPTSGLTPAGGTVSTSNPDA 272
Query: 363 NHEGISSVPNDETMISGPTKITSAPTGPQPHDGGTGTKVNPNSQGAIPQQSRADVAHPSH 422
G +S SG I G G V S A ++ +
Sbjct: 273 AASGGASTATGSKAPSGAVTI---------RVGAQGNNVQSGSSNAQSSAPSGTISSSTS 323
Query: 423 FKSGQHDKLGPTSGSSSLKKTKMKA---RGHQ 451
+G L P +++ + KA RGH+
Sbjct: 324 GGAGTPAALVPMDIDENIEIPESKARVLRGHE 355
>RPL1_HUMAN (Q8IWN7) Retinitis pigmentosa 1-like 1 protein
Length = 2480
Score = 35.0 bits (79), Expect = 0.99
Identities = 33/126 (26%), Positives = 49/126 (38%), Gaps = 7/126 (5%)
Query: 342 DSAGSGISPTKGKVVNTNDEPNHEGISSVPNDETMISGPTKITSAPTGPQPHDGGTGTKV 401
++ G G G V EP G+S + E S P+ TS+ G + H
Sbjct: 592 ETQGQGTEQATGAAVTR--EPLVLGLSCSWDSEGASSTPSTCTSSQQGQRRHRSRASAMS 649
Query: 402 NPNSQGAIPQQSRADVAHPSHFKSGQHDKLGPTSGSSSLKKTKMKARGHQRVEGSSEPRR 461
+P+S G R H SH++ H L SS K+ + + S PR
Sbjct: 650 SPSSPGLGRVAPRGHPRH-SHYRKDTHSPL----DSSVTKQVPRPPERRRACQDGSVPRY 704
Query: 462 SPNASS 467
S ++SS
Sbjct: 705 SGSSSS 710
>PNK1_SCHPO (O13911) Bifunctional polynucleotide phosphatase/kinase
(Polynucleotide kinase-3'-phosphatase) (DNA
5'-kinase/3'-phosphatase) [Includes: Polynucleotide
3'-phosphatase (EC 3.1.3.32) (2'(3')-polynucleotidase);
Polynucleotide 5'-hydroxy-kinase (E
Length = 421
Score = 35.0 bits (79), Expect = 0.99
Identities = 19/60 (31%), Positives = 34/60 (56%), Gaps = 7/60 (11%)
Query: 401 VNPNSQGAIPQQS--RADVAHPSHFKSGQHDKL-----GPTSGSSSLKKTKMKARGHQRV 453
V PN + P+ R +HP HFK +H ++ P+SG S+L ++++ +G++RV
Sbjct: 226 VPPNFESFHPKNYLVRNSSSHPYHFKKSEHQEIVVLVGFPSSGKSTLAESQIVTQGYERV 285
>OMB_DROME (Q24432) Optomotor-blind protein
(Lethal(1)optomotor-blind) (L(1)omb) (Bifid protein)
Length = 972
Score = 35.0 bits (79), Expect = 0.99
Identities = 38/179 (21%), Positives = 67/179 (37%), Gaps = 26/179 (14%)
Query: 261 DSQENCQNKHQAAANQTKHSGGKGAETRFSILEQMDVDAIHGSSGIGASSSGILA*VQIP 320
+S N N + + T ++ A + Q+ + H SS +S+
Sbjct: 59 NSGNNNSNSNNNTNSNTNNTNNLVAVSPTGGGAQLSPQSNHSSSNTTTTSN-----TNNS 113
Query: 321 EQNNLNQNTPTNQESQPQHMVDSAGSGISPTKGKVVNTNDEPNHEGISSVPNDETMISGP 380
NN N N+ N + + ++ + S +G ++T +EP + P P
Sbjct: 114 SSNNNNNNSTHNNNNNHTNNNNNNNNNTSQKQGHHLSTTEEPPSP--AGTP--------P 163
Query: 381 TKITSAPTGPQPHDGGTGTKVNPNSQGAIPQQSRADVAHPSHFKSGQHDKLGPTSGSSS 439
I P P P N NS + S + AHPSH + H P++G+++
Sbjct: 164 PTIVGLPPIPPP---------NNNSSSSSSNNSASAAAHPSHHPTAAHH--SPSTGAAA 211
>PYGO_DROME (Q9V9W8) Pygopus protein (Gammy legs protein)
Length = 815
Score = 34.7 bits (78), Expect = 1.3
Identities = 43/159 (27%), Positives = 52/159 (32%), Gaps = 23/159 (14%)
Query: 320 PEQNNLNQNT-PTNQESQP--------QHMVDSA-GSGISPTKGKVVNTN---DEPNHEG 366
P NN N N P + P QHM G G P G +N +P+H G
Sbjct: 517 PNNNNSNNNNGPMMGQMIPNAVPMQHQQHMGGGPPGHGPGPMPGMGMNQMLPPQQPSHLG 576
Query: 367 ISSVPNDETMISGPTKITSAPTGPQPHD-GGTGTKVNPNSQGAIPQQSRADVAHPSHFKS 425
P M++ P P GP PH GG G P G P H
Sbjct: 577 ----PPHPNMMNHPHHPHHHPGGPPPHMMGGPGMHGGP--AGMPPHMGGGP---NPHMMG 627
Query: 426 GQHDKLGPTSGSSSLKKTKMKARGHQRVEGSSEPRRSPN 464
G H GP G + G + G P SP+
Sbjct: 628 GPHGNAGPHMGHGHMGGVPGPGPGPGGMNGPPHPHMSPH 666
>ACIN_MOUSE (Q9JIX8) Apoptotic chromatin condensation inducer in the
nucleus (Acinus)
Length = 1338
Score = 34.7 bits (78), Expect = 1.3
Identities = 25/108 (23%), Positives = 43/108 (39%), Gaps = 9/108 (8%)
Query: 362 PNHEGISSVPNDETMISGPTKITSAPTGPQPHDGGTGTKVNPNSQGAIPQQSRADVAHPS 421
P H G S + + S P+ +S P P + + +P S+ Q+R
Sbjct: 566 PEHSGKQSADSSSSRSSSPSSSSSPSRSPSPDSVASRPQSSPGSKQRDGAQARV------ 619
Query: 422 HFKSGQHDKLGPTSGSSSLKKTKMKARGHQRVEGSSEPRRSPNASSNA 469
H + K+G S S S +++ ++R SS SP S ++
Sbjct: 620 HANPHERPKMGSRSTSESRSRSRSRSRS---ASSSSRKSLSPGVSRDS 664
>PAK4_MOUSE (Q8BTW9) Serine/threonine-protein kinase PAK 4 (EC
2.7.1.37) (p21-activated kinase 4) (PAK-4)
Length = 593
Score = 34.3 bits (77), Expect = 1.7
Identities = 46/179 (25%), Positives = 70/179 (38%), Gaps = 34/179 (18%)
Query: 339 HMVDSAGSG----ISPTKGKVVNTNDEPNHEGISSVPNDETMISGPTKITSAPTGPQPHD 394
H +GSG + P K + ++ D P G D+ +SGP T PQP
Sbjct: 141 HSEAGSGSGDRRRVGPEK-RPKSSRDGPG--GPQEASRDKRPLSGPDVST-----PQPGS 192
Query: 395 GGTGTKVNPNSQGAIPQQSRADVAHPSHFKSGQHDKLGPT-----------SGSSSLKKT 443
+GTK+ RAD HP G+ + P S SSS T
Sbjct: 193 LTSGTKLAAGRP--FNTYPRADTDHPPRGAQGEPHTMAPNGPSATGLAAPQSSSSSRPPT 250
Query: 444 KMKARGHQRVEG--SSEPRRSPNASS-------NAPGDMGGRTFRERDRSLSPRKHRSS 493
+ + V G +SEP+ +P A + APG G R+ + + +S + R++
Sbjct: 251 RARGAPSPGVLGPHASEPQLAPPARALAAPAVPPAPGPPGPRSPQREPQRVSHEQFRAA 309
>P54_ENTFC (P13692) P54 protein precursor
Length = 516
Score = 34.3 bits (77), Expect = 1.7
Identities = 25/140 (17%), Positives = 56/140 (39%), Gaps = 7/140 (5%)
Query: 258 SMDDSQENCQNKHQAAANQTKHSGGKGAETRFSILEQMDVDAIHGSSGIGASSSGILA*V 317
++ + ++ A ++ + + + T S + + S+G ++ S
Sbjct: 267 ALSSASTTTESSSAAQSSSEESKAPESSTTEESTSTESSTTTENSSTGSSSTESSSTEES 326
Query: 318 QIPEQNNLNQNTPTNQESQPQHMVDSAGSGISPTKGKVVNTNDEPNHEGISSVPNDETMI 377
+PE + ++TP N ES S+ S + +TN+ N+ ++ N+ T+
Sbjct: 327 TVPESST-QESTPANTESS------SSSSNTNVNNNTNNSTNNSTNNSTTNNNNNNNTVT 379
Query: 378 SGPTKITSAPTGPQPHDGGT 397
PT + P P+ G+
Sbjct: 380 PAPTPTPTPAPAPAPNPSGS 399
>HORN_HUMAN (Q86YZ3) Hornerin
Length = 2850
Score = 34.3 bits (77), Expect = 1.7
Identities = 53/245 (21%), Positives = 82/245 (32%), Gaps = 40/245 (16%)
Query: 240 EPGQGN*IQHGGTWRGLWSMDDSQENCQNKHQAAANQTKHSGGKGAETRFSILEQMDVDA 299
E G G G+ G +S Q +H + + Q+ SG + + S A
Sbjct: 821 ESGSGQGYSQHGSASGHFSS-------QGRHGSTSGQSSSSGQHDSSSGQSSSYGQHESA 873
Query: 300 IHGSSGIGASSSGILA*VQIPEQNNLNQNTPTNQESQPQHMVDSAGSGISPTKGKVVNTN 359
H +SG G SG P H +GSG SP+ G+ + +
Sbjct: 874 SHHASGRGRHGSG--------------------SGQSPGHGQRGSGSGQSPSYGRHGSGS 913
Query: 360 DEPNHEGISSVPNDETMISGPTKITSAPTGPQPHDGGTGTKVNPNSQGAIPQQSRADVAH 419
+ G + ++ G + +G H G+G + G+ QS H
Sbjct: 914 GRSSSSGRHGSGSGQSSGFGHKSSSGQSSGYTQHGSGSGHSSSYEQHGSRSGQSSRSEQH 973
Query: 420 PS---------HFKSGQHDKLGPTSGSSSLKKTKMKARG-HQRVEGSSE---PRRSPNAS 466
S SG LG S ++ +RG H G S P RS +
Sbjct: 974 GSSSGSSSSYGQHGSGSRQSLGHGQHGSGSGQSPSPSRGRHGSGSGQSSSYGPYRSGSGW 1033
Query: 467 SNAPG 471
S++ G
Sbjct: 1034 SSSRG 1038
>PNT2_DROME (P51023) ETS-like protein pointed, isoform P2 (D-ETS-2)
Length = 718
Score = 33.9 bits (76), Expect = 2.2
Identities = 50/236 (21%), Positives = 92/236 (38%), Gaps = 30/236 (12%)
Query: 258 SMD--DSQENCQNKHQAAANQTKHSGGKGAETRFSI-------LEQMDVDAIHGSSGIGA 308
SMD S N NK+ A H+G + T I +Q ++GS +
Sbjct: 299 SMDYVTSAGNSDNKNFHRATPHSHNGYNTSNTHDRINNSTPPQQQQSQQPTVNGSGSASS 358
Query: 309 SSSGILA*VQIPEQNNLNQNTPTNQESQPQHMVDSAGSGISPTKGKVVNTNDEPNHEGIS 368
+++ + + + NN N NT ++ + + +++G + G N N+ N ++
Sbjct: 359 NNNNSMLPPAVQQSNNENNNTSSSNTNNSSNNNNNSGGSNNSNAGSNNNNNNNNNINFMA 418
Query: 369 SVPNDETMISGPTKITSAPTGPQPHDGGTGTKVNPN--SQGAIPQQSRADVAHPSHFKSG 426
+ + + + G + GG+ ++ +P S +P AH + + G
Sbjct: 419 AAAIFQHHLKEEPGTQNGNIG--GYGGGSNSQNDPTDLSSYGLP-------AHLAAYGGG 469
Query: 427 QHDKLGPTSGSS-------SLKKTKMKARGHQRVEGSSEPRRSPNASSNAPGDMGG 475
GPT G S S + + A+ HQ + SS S AS + G+ G
Sbjct: 470 SGS--GPTGGRSSGGGGDESDYHSTISAQDHQS-QQSSGGNGSGGASGGSTGNSNG 522
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.328 0.141 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 124,007,474
Number of Sequences: 164201
Number of extensions: 5491311
Number of successful extensions: 15525
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 58
Number of HSP's that attempted gapping in prelim test: 15445
Number of HSP's gapped (non-prelim): 132
length of query: 992
length of database: 59,974,054
effective HSP length: 120
effective length of query: 872
effective length of database: 40,269,934
effective search space: 35115382448
effective search space used: 35115382448
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0113.5


