

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
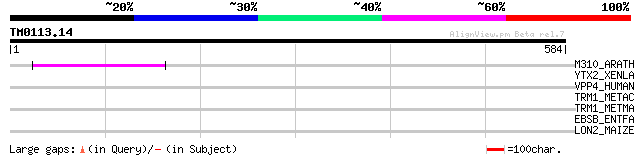
Query= TM0113.14
(584 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 107 9e-23
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 38 0.064
VPP4_HUMAN (Q9HBG4) Vacuolar proton translocating ATPase 116 kDa... 33 1.6
TRM1_METAC (Q8TGX6) N(2),N(2)-dimethylguanosine tRNA methyltrans... 33 2.1
TRM1_METMA (Q8PU28) N(2),N(2)-dimethylguanosine tRNA methyltrans... 33 2.7
EBSB_ENTFA (P36921) Cell wall enzyme ebsB 33 2.7
LON2_MAIZE (P93648) Lon protease homolog 2, mitochondrial precur... 31 7.8
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 107 bits (267), Expect = 9e-23
Identities = 54/142 (38%), Positives = 79/142 (55%), Gaps = 2/142 (1%)
Query: 25 LPTYAMAILKFPKQFCHQIASQIARFWWRG-NNDRGIHWKNWASLMFSKAE-GGMGFKDF 82
LP YAM+ + K C ++ S + FWW N R I W W L SK + GG+GF+D
Sbjct: 3 LPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRDL 62
Query: 83 AVLNQACLAKQAWRILQEPNAFWASVLKGIYFPNENFLHAKKKRQASWAWNSIIHGRELI 142
NQA LAKQ++RI+ +P+ + +L+ YFP+ + + + S+AW SIIHGREL+
Sbjct: 63 GWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGRELL 122
Query: 143 LNQGRWSIEDGKSVNLLGHRWL 164
+I DG + RW+
Sbjct: 123 SRGLLRTIGDGIHTKVWLDRWI 144
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein (ORF
2)
Length = 1308
Score = 38.1 bits (87), Expect = 0.064
Identities = 22/72 (30%), Positives = 33/72 (45%), Gaps = 2/72 (2%)
Query: 255 SEPSKECWRSI*SARIPHKIKVFLWKIMHEALPTKVNLRRRHCVSETSCPICEAPREDAA 314
+E + WR+ S+ +P WK++H AL T L R S +CP C E
Sbjct: 1115 NEGERPQWRAFYSSLVPRPTGDLSWKVLHGALSTGEYL-ARFTDSPAACPFC-GKGESVF 1172
Query: 315 HVFLHCPWLRPV 326
H + C L+P+
Sbjct: 1173 HAYFTCARLQPL 1184
>VPP4_HUMAN (Q9HBG4) Vacuolar proton translocating ATPase 116 kDa
subunit a isoform 4 (V-ATPase 116-kDa isoform a4)
(Vacuolar proton translocating ATPase 116 kDa subunit a
kidney isoform)
Length = 840
Score = 33.5 bits (75), Expect = 1.6
Identities = 21/56 (37%), Positives = 27/56 (47%), Gaps = 9/56 (16%)
Query: 132 WNSIIHGRELILNQGRWSIEDG--------KSVNLLGHRWLACNERLREGPLDTTV 179
WN+ HGR LIL G +SI G KS+N+ G W + R G +T V
Sbjct: 442 WNTFFHGRYLILLMGIFSIYTGLIYNDCFSKSLNIFGSSW-SVQPMFRNGTWNTHV 496
>TRM1_METAC (Q8TGX6) N(2),N(2)-dimethylguanosine tRNA
methyltransferase (EC 2.1.1.32)
(tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA
2,2-dimethylguanosine-26 methyltransferase)
(tRNA(m(2,2)G26)dimethyltransferase)
Length = 388
Score = 33.1 bits (74), Expect = 2.1
Identities = 24/72 (33%), Positives = 36/72 (49%), Gaps = 2/72 (2%)
Query: 391 PVLCLNLAVEASKEFLLETTTEFLPIYGGHVNRGIRHWTRPPLGFLKCNTDAGFLRVGNK 450
P L+ A A++ L T T+ P+ G H+N GIR + PL + +++ G LRV
Sbjct: 143 PAPYLDAAATAAQGMLSVTATDTAPLCGAHLNSGIRKYAAVPLN-TEYHSEMG-LRVLLG 200
Query: 451 AAGGYIFKDDKG 462
A K +KG
Sbjct: 201 ACARECAKHEKG 212
>TRM1_METMA (Q8PU28) N(2),N(2)-dimethylguanosine tRNA
methyltransferase (EC 2.1.1.32)
(tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA
2,2-dimethylguanosine-26 methyltransferase)
(tRNA(m(2,2)G26)dimethyltransferase)
Length = 388
Score = 32.7 bits (73), Expect = 2.7
Identities = 24/74 (32%), Positives = 38/74 (50%), Gaps = 4/74 (5%)
Query: 389 PNPVLCLNLAVEASKEFLLETTTEFLPIYGGHVNRGIRHWTRPPLGFLKCNTDAGFLRVG 448
P+P L + A ++ L T T+ P+ G H+N GIR + PL + +++ G LRV
Sbjct: 143 PSPYL--DAAASSAYSMLSVTATDTAPLCGAHLNSGIRKYASVPLN-TEYHSEMG-LRVL 198
Query: 449 NKAAGGYIFKDDKG 462
A + K +KG
Sbjct: 199 LGACARELAKHEKG 212
>EBSB_ENTFA (P36921) Cell wall enzyme ebsB
Length = 135
Score = 32.7 bits (73), Expect = 2.7
Identities = 31/137 (22%), Positives = 60/137 (43%), Gaps = 13/137 (9%)
Query: 436 LKCNTDAGFLRVGNKAAGGYIFKDDKGCFV*GNHRQLHAASALA----AEALILREAVFA 491
L+ DA ++ GG ++ D+ + +QLH + AE +L EA+
Sbjct: 2 LRIYVDAATKGNPGESGGGIVYLTDQ------SRQQLHVPLGIVSNHEAEFKVLIEALKK 55
Query: 492 AM--NLNMDRVVFEADNLGLIEACREQREKGE-ILAIVKDIRKCKLDFPNWIFSWTRREG 548
A+ N V+ +D+ +++ + K E + + ++ + +FP + W
Sbjct: 56 AIANEDNQQTVLLHSDSKIVVQTIEKNYAKNEKYQPYLAEYQQLEKNFPLLLIEWLPESQ 115
Query: 549 NECAHVLARLALCRSIP 565
N+ A +LAR AL + P
Sbjct: 116 NKAADMLARQALQKFYP 132
>LON2_MAIZE (P93648) Lon protease homolog 2, mitochondrial precursor
(EC 3.4.21.-)
Length = 964
Score = 31.2 bits (69), Expect = 7.8
Identities = 22/59 (37%), Positives = 27/59 (45%), Gaps = 1/59 (1%)
Query: 153 GKSVNLLGHRWLACNERLREGPLDTTVSSLMEN-QTRTWSISKITSVLSLPDLGRVLQT 210
G V LLGHR L E + E PL V L E + + K TS + L VL+T
Sbjct: 180 GDHVVLLGHRRLRITEMVEEDPLTVKVDHLKEKPYNKDDDVMKATSFEVISTLREVLRT 238
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.138 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 72,177,578
Number of Sequences: 164201
Number of extensions: 3075063
Number of successful extensions: 7086
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 7083
Number of HSP's gapped (non-prelim): 7
length of query: 584
length of database: 59,974,054
effective HSP length: 116
effective length of query: 468
effective length of database: 40,926,738
effective search space: 19153713384
effective search space used: 19153713384
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0113.14


