

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
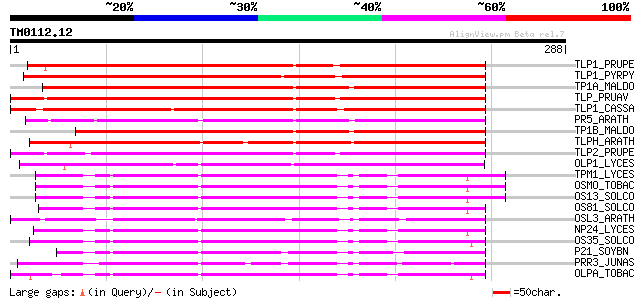
Query= TM0112.12
(288 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TLP1_PRUPE (P83332) Thaumatin-like protein 1 precursor (PpAZ44) 246 6e-65
TLP1_PYRPY (O80327) Thaumatin-like protein 1 precursor 245 8e-65
TP1A_MALDO (Q9FSG7) Thaumatin-like protein 1a precursor (Allerge... 244 2e-64
TLP_PRUAV (P50694) Thaumatin-like protein precursor 240 3e-63
TLP1_CASSA (Q9SMH2) Thaumatin-like protein 1 precursor 237 2e-62
PR5_ARATH (P28493) Pathogenesis-related protein 5 precursor (PR-5) 233 5e-61
TP1B_MALDO (P83336) Thaumatin-like protein 1b (Pathogenesis-rela... 232 7e-61
TLPH_ARATH (P50699) Thaumatin-like protein precursor 230 3e-60
TLP2_PRUPE (P83335) Thaumatin-like protein 2 precursor (PpAZ8) 219 8e-57
OLP1_LYCES (Q41350) Osmotin-like protein precursor 197 3e-50
TPM1_LYCES (Q01591) Osmotin-like protein TPM-1 precursor (PR P23... 182 6e-46
OSMO_TOBAC (P14170) Osmotin precursor 179 9e-45
OS13_SOLCO (P50701) Osmotin-like protein OSML13 precursor (PA13) 177 3e-44
OS81_SOLCO (P50702) Osmotin-like protein OSML81 precursor (PA81) 175 1e-43
OSL3_ARATH (P50700) Osmotin-like protein OSM34 precursor 172 9e-43
NP24_LYCES (P12670) NP24 protein precursor (Pathogenesis-related... 172 9e-43
OS35_SOLCO (P50703) Osmotin-like protein OSML15 precursor (PA15) 166 6e-41
P21_SOYBN (P25096) P21 protein 163 4e-40
PRR3_JUNAS (P81295) Pathogenesis-related protein precursor (Poll... 163 5e-40
OLPA_TOBAC (P25871) Osmotin-like protein precursor (Pathogenesis... 163 5e-40
>TLP1_PRUPE (P83332) Thaumatin-like protein 1 precursor (PpAZ44)
Length = 246
Score = 246 bits (627), Expect = 6e-65
Identities = 127/243 (52%), Positives = 153/243 (62%), Gaps = 9/243 (3%)
Query: 10 LLSLFTLS--FLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHF 67
LL L TL+ F S +A NKC +T+WPG L+G P L TGF L +G SR++
Sbjct: 8 LLGLTTLAILFFSGAHAAKITFTNKCSYTVWPGTLTGDQKPQLSLTGFELATGISRSVDA 67
Query: 68 PKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADGLDF 126
P WSGR + RT C D GKF+C TADCGS +V C G GA PPATL E T+ G DF
Sbjct: 68 PSPWSGRFFGRTRCSTDASGKFTCATADCGSGQVSCNGNGAAPPATLVEITIASNGGQDF 127
Query: 127 YDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSV-ACMSA 185
YDVSLVDG+NLPM + + GT G C + C D+N CP L +V G+ GSV AC SA
Sbjct: 128 YDVSLVDGFNLPMSVAPQGGT-GKCKASTCPADINKVCPAPL---QVKGSDGSVIACKSA 183
Query: 186 CEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCAS-ANYLII 244
C AF P+YCC+ P+TC P YS FK CP+AYSYAYDDK+ST+TC+ YLI
Sbjct: 184 CLAFNQPKYCCTPPNDKPETCPPPDYSKLFKTQCPQAYSYAYDDKSSTFTCSGRPAYLIT 243
Query: 245 FCP 247
FCP
Sbjct: 244 FCP 246
>TLP1_PYRPY (O80327) Thaumatin-like protein 1 precursor
Length = 244
Score = 245 bits (626), Expect = 8e-65
Identities = 127/243 (52%), Positives = 153/243 (62%), Gaps = 7/243 (2%)
Query: 8 LILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHF 67
LI L L LS + V SA F NKC +T+WPG L+G P L +TGF L SG S +L
Sbjct: 6 LIGLVLVFLSEHAGVYSAKFTFTNKCPNTVWPGTLTGGGGPQLLSTGFELASGASTSLTV 65
Query: 68 PKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADGLDF 126
WSGR W R+ C D GKF C T DCGS ++ C G GA PPA+L E TL G DF
Sbjct: 66 QAPWSGRFWGRSHCSIDSSGKFKCSTGDCGSGQISCNGAGASPPASLVELTLATNGGQDF 125
Query: 127 YDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSV-ACMSA 185
YDVSLVDG+NLP+ + A RG G C+ T C ++N CP +L G+ GSV C SA
Sbjct: 126 YDVSLVDGFNLPIKL-APRGGSGDCNSTSCAANINTVCPAELS---DKGSDGSVIGCKSA 181
Query: 186 CEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC-ASANYLII 244
C A P+YCC+ AY TPDTC P+ +S FK+ CP+AYSYAYDDK+ST+TC NY I
Sbjct: 182 CLALNQPQYCCTGAYGTPDTCPPTDFSKVFKNQCPQAYSYAYDDKSSTFTCFGGPNYEIT 241
Query: 245 FCP 247
FCP
Sbjct: 242 FCP 244
>TP1A_MALDO (Q9FSG7) Thaumatin-like protein 1a precursor (Allergen
Mal d 2) (Mdtl1) (Pathogenesis-related protein 5a)
(PR-5a)
Length = 246
Score = 244 bits (622), Expect = 2e-64
Identities = 117/232 (50%), Positives = 149/232 (63%), Gaps = 5/232 (2%)
Query: 18 FLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWSGRIWA 77
F S +A N C +T+WPG L+G P L TGF L S SR++ P WSGR W
Sbjct: 18 FFSGAHAAKITFTNNCPNTVWPGTLTGDQKPQLSLTGFELASKASRSVDAPSPWSGRFWG 77
Query: 78 RTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADGLDFYDVSLVDGYN 136
RT C D GKF+C TADCGS +V C G GA PPATL E T+ G D+YDVSLVDG+N
Sbjct: 78 RTRCSTDAAGKFTCETADCGSGQVACNGAGAVPPATLVEITIAANGGQDYYDVSLVDGFN 137
Query: 137 LPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDPRYCC 196
LPM + + GT G C P+ C ++N CP L+V +G+ ++C SAC AFGD +YCC
Sbjct: 138 LPMSVAPQGGT-GECKPSSCPANVNKVCPAPLQVKAADGS--VISCKSACLAFGDSKYCC 194
Query: 197 SEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASA-NYLIIFCP 247
+ +TP+TC P+ YS F+ CP+AYSYAYDDK ST+TC+ +Y+I FCP
Sbjct: 195 TPPNNTPETCPPTEYSEIFEKQCPQAYSYAYDDKNSTFTCSGGPDYVITFCP 246
>TLP_PRUAV (P50694) Thaumatin-like protein precursor
Length = 245
Score = 240 bits (612), Expect = 3e-63
Identities = 123/249 (49%), Positives = 151/249 (60%), Gaps = 6/249 (2%)
Query: 1 MGRLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESG 60
M + L+ ++ LSL LSF +A+ N C + +WPG L+ P L TTGF L S
Sbjct: 1 MMKTLVVVLSLSLTILSF-GGAHAATISFKNNCPYMVWPGTLTSDQKPQLSTTGFELASQ 59
Query: 61 KSRTLHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLN 119
S L P W+GR WART C D GKF C TADC S +V C G GA PPATLAEF +
Sbjct: 60 ASFQLDTPVPWNGRFWARTGCSTDASGKFVCATADCASGQVMCNGNGAIPPATLAEFNIP 119
Query: 120 GADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGS 179
G DFYDVSLVDG+NLPM + + GT G C C ++N CP +L+ + G
Sbjct: 120 AGGGQDFYDVSLVDGFNLPMSVTPQGGT-GDCKTASCPANVNAVCPSELQ--KKGSDGSV 176
Query: 180 VACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC-AS 238
VAC+SAC FG P+YCC+ +TP+TC P+ YS F +ACP AYSYAYDDK T+TC
Sbjct: 177 VACLSACVKFGTPQYCCTPPQNTPETCPPTNYSEIFHNACPDAYSYAYDDKRGTFTCNGG 236
Query: 239 ANYLIIFCP 247
NY I FCP
Sbjct: 237 PNYAITFCP 245
>TLP1_CASSA (Q9SMH2) Thaumatin-like protein 1 precursor
Length = 243
Score = 237 bits (605), Expect = 2e-62
Identities = 126/250 (50%), Positives = 153/250 (60%), Gaps = 11/250 (4%)
Query: 1 MGRLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESG 60
M L L+ + L+LF FLS SA N C TIWPG L+ P LP TGF L S
Sbjct: 2 MKTLALYGLTLALF---FLSGAHSAKITFTNNCPRTIWPGTLTSDQKPQLPNTGFVLASK 58
Query: 61 KSRTLHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLN 119
S TL W GR WART C + GKF+C TADC + +V C G GA PPA+L E +
Sbjct: 59 ASLTLGVQAPWKGRFWARTRCTTN-SGKFTCETADCSTGQVACNGNGAIPPASLVEINIA 117
Query: 120 GADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGS 179
G+DFYDVSLVDGYNLP+ + R GT G C T C ++N CP +L+V G+ S
Sbjct: 118 ANRGMDFYDVSLVDGYNLPVSVATRGGT-GDCKATSCRANVNAVCPAELQV---KGSDAS 173
Query: 180 V-ACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCAS 238
V AC SAC AF P+YCC+ A+ T TC + YS FK CP+AYSYAYDD TST+TC+
Sbjct: 174 VLACKSACTAFNQPQYCCTGAFDTARTCPATKYSRIFKQQCPQAYSYAYDDSTSTFTCSG 233
Query: 239 A-NYLIIFCP 247
A +Y+I FCP
Sbjct: 234 APDYVITFCP 243
>PR5_ARATH (P28493) Pathogenesis-related protein 5 precursor (PR-5)
Length = 239
Score = 233 bits (593), Expect = 5e-61
Identities = 124/239 (51%), Positives = 142/239 (58%), Gaps = 8/239 (3%)
Query: 9 ILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFP 68
IL +F S ++ V + F + N C T+W G L+G P L GF L G SR L P
Sbjct: 9 ILFLVFITSGIA-VMATDFTLRNNCPTTVWAGTLAG-QGPKLGDGGFELTPGASRQLTAP 66
Query: 69 KAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYD 128
WSGR WART C D G CVT DCG + C GG PP TLAEFTL G G DFYD
Sbjct: 67 AGWSGRFWARTGCNFDASGNGRCVTGDCGG--LRCNGGGVPPVTLAEFTLVGDGGKDFYD 124
Query: 129 VSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEA 188
VSLVDGYN+ + I G+ G C GC+ DLN CP LKV N VAC SACE
Sbjct: 125 VSLVDGYNVKLGIRPSGGS-GDCKYAGCVSDLNAACPDMLKVMDQNNV---VACKSACER 180
Query: 189 FGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLIIFCP 247
F +YCC A P+TC P+ YS FK+ACP AYSYAYDD+TST+TC ANY I FCP
Sbjct: 181 FNTDQYCCRGANDKPETCPPTDYSRIFKNACPDAYSYAYDDETSTFTCTGANYEITFCP 239
>TP1B_MALDO (P83336) Thaumatin-like protein 1b (Pathogenesis-related
protein 5b) (PR-5b) (Fragment)
Length = 212
Score = 232 bits (592), Expect = 7e-61
Identities = 111/215 (51%), Positives = 141/215 (64%), Gaps = 5/215 (2%)
Query: 35 HTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWSGRIWARTLCGHDRDGKFSCVTA 94
+T+WPG L+G P L T F L S S+++ P WSGR W RT C D GKFSC TA
Sbjct: 1 NTVWPGTLTGDQKPQLSLTAFELASKASQSVDAPSPWSGRFWGRTRCSTDAAGKFSCETA 60
Query: 95 DCGSDKVECIG-GAKPPATLAEFTLNGADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSP 153
DCGS +V C G GA PPATL E T+ G D+YDVSLVDG+NLPM + + GT G C P
Sbjct: 61 DCGSGQVACNGAGAVPPATLVEITIAANGGQDYYDVSLVDGFNLPMSVAPQGGT-GECKP 119
Query: 154 TGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSL 213
+ C ++N CP L+V +G+ ++C SAC AFGD +YCC+ TP+TC P+ YS
Sbjct: 120 SSCPANVNMACPAQLQVKAADGS--VISCKSACLAFGDSKYCCTPPNDTPETCPPTEYSE 177
Query: 214 FFKHACPRAYSYAYDDKTSTYTCASA-NYLIIFCP 247
F+ CP+AYSYAYDDK ST+TC+ +Y+I FCP
Sbjct: 178 IFEKQCPQAYSYAYDDKNSTFTCSGGPDYVITFCP 212
>TLPH_ARATH (P50699) Thaumatin-like protein precursor
Length = 243
Score = 230 bits (586), Expect = 3e-60
Identities = 120/241 (49%), Positives = 147/241 (60%), Gaps = 9/241 (3%)
Query: 11 LSLFTLSFLSEVQSASFEIV---NKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHF 67
++LF +FL + AS V NKC+H +WPG+ A L GF L + K+ +L
Sbjct: 4 INLFLFAFLLLLSHASASTVIFYNKCKHPVWPGIQPSAGQNLLAGGGFKLPANKAHSLQL 63
Query: 68 PKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLNGADGLDF 126
P WSGR W R C DR G+ C T DCG + C G G +PPATLAE TL LDF
Sbjct: 64 PPLWSGRFWGRHGCTFDRSGRGHCATGDCGGS-LSCNGAGGEPPATLAEITLGPE--LDF 120
Query: 127 YDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSAC 186
YDVSLVDGYNL M I+ +G+ G CS GC+ DLN CP L+V NG VAC SAC
Sbjct: 121 YDVSLVDGYNLAMSIMPVKGS-GQCSYAGCVSDLNQMCPVGLQVRSRNGKR-VVACKSAC 178
Query: 187 EAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLIIFC 246
AF P+YCC+ + P +C P+ YS FK ACP+AYSYAYDD TS TC+ ANY++ FC
Sbjct: 179 SAFNSPQYCCTGLFGNPQSCKPTAYSKIFKVACPKAYSYAYDDPTSIATCSKANYIVTFC 238
Query: 247 P 247
P
Sbjct: 239 P 239
>TLP2_PRUPE (P83335) Thaumatin-like protein 2 precursor (PpAZ8)
Length = 242
Score = 219 bits (557), Expect = 8e-57
Identities = 116/249 (46%), Positives = 146/249 (58%), Gaps = 9/249 (3%)
Query: 1 MGRLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESG 60
M + L ++ LSL LSF +A+ N C +T+WP + +P L TTGF L S
Sbjct: 1 MMKTLGAVLSLSLTLLSF-GGAHAATMSFKNNCPYTVWP---ASFGNPQLSTTGFELASQ 56
Query: 61 KSRTLHFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAK-PPATLAEFTLN 119
S L P WSGR WART C D GKF C TADC S ++ C G PPATLAEFT+
Sbjct: 57 ASFQLDTPVPWSGRFWARTRCSTDASGKFVCETADCDSGQLMCNGKTGIPPATLAEFTIA 116
Query: 120 GADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGS 179
G DFYDVSLVDG+NLPM + + GT G C C ++N CP +L+ ++ G
Sbjct: 117 AGGGQDFYDVSLVDGFNLPMSVTPQGGT-GTCKMGSCAANVNLVCPSELQ--KIGSDGSV 173
Query: 180 VACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASA 239
VAC+SAC FG+P+YCC+ T + C P+ YS F CP AYSYA+DD +TC+
Sbjct: 174 VACLSACVKFGEPQYCCTPPQETKEKCPPTNYSQIFHEQCPDAYSYAFDDNKGLFTCSGG 233
Query: 240 -NYLIIFCP 247
NYLI FCP
Sbjct: 234 PNYLITFCP 242
>OLP1_LYCES (Q41350) Osmotin-like protein precursor
Length = 252
Score = 197 bits (500), Expect = 3e-50
Identities = 110/247 (44%), Positives = 140/247 (56%), Gaps = 9/247 (3%)
Query: 6 LFLILLSLFTLSFLSEVQSASF--EIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSR 63
+ L L LFTL LS+ + +F +VN C +TIWP + A P L GF L S R
Sbjct: 8 ILLPLSLLFTLLSLSQSTNPNFILTLVNNCPYTIWPAIQPNAGHPVLERGGFTLHSLTHR 67
Query: 64 TLHFPKA-WSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLN-G 120
+ P A WSGRIWART C + GKF C T DCG ++EC G G PATLA+F L+ G
Sbjct: 68 SFPAPNAHWSGRIWARTGCNYQH-GKFYCATGDCGG-RIECDGLGGAAPATLAQFVLHHG 125
Query: 121 ADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSV 180
Y VSLVDG+N+P+ + G +G C GC +L CP L+ G G V
Sbjct: 126 HADFSTYGVSLVDGFNIPLTVTPHEG-KGVCPVVGCRANLLESCPAVLQFRSHGGHGPVV 184
Query: 181 ACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASAN 240
C SACEAF +CC Y++P TC PS YS FFKHACP ++YA+D + + C+S
Sbjct: 185 GCKSACEAFKSDEFCCRNHYNSPQTCKPSSYSQFFKHACPATFTYAHDSPSLMHECSSPR 244
Query: 241 YL-IIFC 246
L +IFC
Sbjct: 245 ELKVIFC 251
>TPM1_LYCES (Q01591) Osmotin-like protein TPM-1 precursor (PR P23)
(Fragment)
Length = 238
Score = 182 bits (463), Expect = 6e-46
Identities = 100/247 (40%), Positives = 136/247 (54%), Gaps = 24/247 (9%)
Query: 14 FTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWS- 72
F L+F++ +A+FE+ N C +T+W A S P+ G L+ G++ ++ P+
Sbjct: 3 FLLAFVTYTYAATFEVRNNCPYTVW------AASTPIGG-GRRLDRGQTWVINAPRGTKM 55
Query: 73 GRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYDVSLV 132
RIW RT C D DG+ SC T DCG ++C G KPP TLAE+ L+ LDF+D+SLV
Sbjct: 56 ARIWGRTNCNFDGDGRGSCQTGDCGG-VLQCTGWGKPPNTLAEYALDQFSNLDFWDISLV 114
Query: 133 DGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDP 192
DG+N+PM + G C C ++NG CP L+V GG C + C FG
Sbjct: 115 DGFNIPMTFAPTNPSGGKCHAIHCTANINGECPGSLRV-----PGG---CNNPCTTFGGQ 166
Query: 193 RYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC--ASANYLIIFCPLPY 250
+YCC T C P+ S FFK CP AYSY DD TST+TC S NY ++FCP
Sbjct: 167 QYCC-----TQGPCGPTDLSRFFKQRCPDAYSYPQDDPTSTFTCPSGSTNYRVVFCPNGV 221
Query: 251 TRYNYSI 257
T N+ +
Sbjct: 222 TSPNFPL 228
>OSMO_TOBAC (P14170) Osmotin precursor
Length = 246
Score = 179 bits (453), Expect = 9e-45
Identities = 96/247 (38%), Positives = 136/247 (54%), Gaps = 24/247 (9%)
Query: 14 FTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWS- 72
F L+ ++ +A+ E+ N C +T+W A S P+ G L+ G++ ++ P+
Sbjct: 11 FLLALVTYTYAATIEVRNNCPYTVW------AASTPIGG-GRRLDRGQTWVINAPRGTKM 63
Query: 73 GRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYDVSLV 132
R+W RT C + G+ +C T DCG ++C G KPP TLAE+ L+ GLDF+D+SLV
Sbjct: 64 ARVWGRTNCNFNAAGRGTCQTGDCGG-VLQCTGWGKPPNTLAEYALDQFSGLDFWDISLV 122
Query: 133 DGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDP 192
DG+N+PM + G C C ++NG CPR+L+V GG C + C FG
Sbjct: 123 DGFNIPMTFAPTNPSGGKCHAIHCTANINGECPRELRV-----PGG---CNNPCTTFGGQ 174
Query: 193 RYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC--ASANYLIIFCPLPY 250
+YCC T C P+ +S FFK CP AYSY DD TST+TC S NY +IFCP
Sbjct: 175 QYCC-----TQGPCGPTFFSKFFKQRCPDAYSYPQDDPTSTFTCPGGSTNYRVIFCPNGQ 229
Query: 251 TRYNYSI 257
N+ +
Sbjct: 230 AHPNFPL 236
>OS13_SOLCO (P50701) Osmotin-like protein OSML13 precursor (PA13)
Length = 246
Score = 177 bits (449), Expect = 3e-44
Identities = 98/247 (39%), Positives = 134/247 (53%), Gaps = 24/247 (9%)
Query: 14 FTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWS- 72
F L+F++ +A+ E+ N C +T+W A S P+ G L+ G++ ++ P+
Sbjct: 11 FLLAFVTYTYAATIEVRNNCPYTVW------AASTPIGG-GRRLDRGQTWVINAPRGTKM 63
Query: 73 GRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYDVSLV 132
RIW RT C D G+ SC T DCG ++C G KPP TLAE+ L+ LDF+D+SLV
Sbjct: 64 ARIWGRTNCNFDGAGRGSCQTGDCGG-VLQCTGWGKPPNTLAEYALDQFSNLDFWDISLV 122
Query: 133 DGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDP 192
DG+N+PM + G C C ++NG CP L+V GG C + C FG
Sbjct: 123 DGFNIPMTFAPTNPSGGKCHAIHCTANINGECPGSLRV-----PGG---CNNPCTTFGGQ 174
Query: 193 RYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC--ASANYLIIFCPLPY 250
+YCC T C P+ S FFK CP AYSY DD TST+TC S NY ++FCP
Sbjct: 175 QYCC-----TQGPCGPTDLSRFFKQRCPDAYSYPQDDPTSTFTCPSGSTNYRVVFCPNGV 229
Query: 251 TRYNYSI 257
T N+ +
Sbjct: 230 TSPNFPL 236
>OS81_SOLCO (P50702) Osmotin-like protein OSML81 precursor (PA81)
Length = 247
Score = 175 bits (444), Expect = 1e-43
Identities = 96/235 (40%), Positives = 130/235 (54%), Gaps = 24/235 (10%)
Query: 16 LSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWS-GR 74
L+F++ +A+ E+ N C +T+W A S P+ G L G++ ++ P+ R
Sbjct: 13 LAFVTYTYAATIEVRNNCPYTVW------AASTPIGG-GRRLNKGQTWVINAPRGTKMAR 65
Query: 75 IWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYDVSLVDG 134
IW RT C + G+ SC T DCG ++C G KPP TLAE+ L+ LDF+D+SLVDG
Sbjct: 66 IWGRTGCNFNAAGRGSCQTGDCGG-VLQCTGWGKPPNTLAEYALDQFSNLDFWDISLVDG 124
Query: 135 YNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDPRY 194
+N+PM + + G C C ++NG CPR LKV GG C + C FG +Y
Sbjct: 125 FNIPMTFAPTKPSAGKCHAIHCTANINGECPRALKV-----PGG---CNNPCTTFGGQQY 176
Query: 195 CCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC--ASANYLIIFCP 247
CC T C P+ S FFK CP AYSY DD TST+TC S NY ++FCP
Sbjct: 177 CC-----TQGPCGPTELSKFFKKRCPDAYSYPQDDPTSTFTCPSGSTNYRVVFCP 226
>OSL3_ARATH (P50700) Osmotin-like protein OSM34 precursor
Length = 244
Score = 172 bits (436), Expect = 9e-43
Identities = 97/248 (39%), Positives = 130/248 (52%), Gaps = 26/248 (10%)
Query: 1 MGRLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESG 60
M LL+ + S L +S +A+FEI+N+C +T+W G G L++G
Sbjct: 1 MANLLVSTFIFSALLL--ISTATAATFEILNQCSYTVWAAASPGG--------GRRLDAG 50
Query: 61 KSRTLHFPKAWS-GRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLN 119
+S L RIW RT C D G+ C T DC S ++C G +PP TLAE+ LN
Sbjct: 51 QSWRLDVAAGTKMARIWGRTNCNFDSSGRGRCQTGDC-SGGLQCTGWGQPPNTLAEYALN 109
Query: 120 GADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGS 179
+ LDFYD+SLVDG+N+PM T C C D+NG CP L+ GG
Sbjct: 110 QFNNLDFYDISLVDGFNIPMEFSP---TSSNCHRILCTADINGQCPNVLRAP-----GG- 160
Query: 180 VACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASA 239
C + C F +YCC+ + C+ + YS FFK CP AYSY DD TST+TC +
Sbjct: 161 --CNNPCTVFQTNQYCCTNGQGS---CSDTEYSRFFKQRCPDAYSYPQDDPTSTFTCTNT 215
Query: 240 NYLIIFCP 247
NY ++FCP
Sbjct: 216 NYRVVFCP 223
>NP24_LYCES (P12670) NP24 protein precursor (Pathogenesis-related
protein PR P23) (Salt-induced protein)
Length = 247
Score = 172 bits (436), Expect = 9e-43
Identities = 96/238 (40%), Positives = 130/238 (54%), Gaps = 24/238 (10%)
Query: 13 LFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWS 72
LF L ++ +A+ E+ N C +T+W A S P+ G L G++ ++ P+
Sbjct: 10 LFFLLCVTYTYAATIEVRNNCPYTVW------AASTPIGG-GRRLNRGQTWVINAPRGTK 62
Query: 73 -GRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYDVSL 131
RIW RT C + G+ +C T DCG ++C G KPP TLAE+ L+ LDF+D+SL
Sbjct: 63 MARIWGRTGCNFNAAGRGTCQTGDCGG-VLQCTGWGKPPNTLAEYALDQFSNLDFWDISL 121
Query: 132 VDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGD 191
VDG+N+PM + + G C C ++NG CPR LKV GG C + C FG
Sbjct: 122 VDGFNIPMTFAPTKPSGGKCHAIHCTANINGECPRALKV-----PGG---CNNPCTTFGG 173
Query: 192 PRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC--ASANYLIIFCP 247
+YCC T C P+ S FFK CP AYSY DD TST+TC S NY ++FCP
Sbjct: 174 QQYCC-----TQGPCGPTELSKFFKKRCPDAYSYPQDDPTSTFTCPGGSTNYRVVFCP 226
>OS35_SOLCO (P50703) Osmotin-like protein OSML15 precursor (PA15)
Length = 250
Score = 166 bits (420), Expect = 6e-41
Identities = 97/241 (40%), Positives = 128/241 (52%), Gaps = 25/241 (10%)
Query: 11 LSLFTLSFLSEVQSAS-FEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPK 69
L F L+F++ ++ FE+ N C +T+W A + P+ G LE G+S P
Sbjct: 8 LVFFLLAFVTYTNASGVFEVHNNCPYTVW------AAATPIGG-GRRLERGQSWWFWAPP 60
Query: 70 AWS-GRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYD 128
RIW RT C D G+ C T DCG +EC G KPP TLAE+ LN LDF+D
Sbjct: 61 GTKMARIWGRTNCNFDGAGRGWCQTGDCGG-VLECKGWGKPPNTLAEYALNQFSNLDFWD 119
Query: 129 VSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEA 188
+S++DG+N+PM G C P C+ ++NG CP L+V GG C + C
Sbjct: 120 ISVIDGFNIPMSFGPTNPGPGKCHPIQCVANINGECPGSLRV-----PGG---CNNPCTT 171
Query: 189 FGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCAS--ANYLIIFC 246
FG +YCC T C P+ S FFK CP AYSY DD TST+TC S +Y ++FC
Sbjct: 172 FGGQQYCC-----TQGPCGPTDLSRFFKQRCPDAYSYPQDDPTSTFTCQSWTTDYKVMFC 226
Query: 247 P 247
P
Sbjct: 227 P 227
>P21_SOYBN (P25096) P21 protein
Length = 202
Score = 163 bits (413), Expect = 4e-40
Identities = 94/226 (41%), Positives = 123/226 (53%), Gaps = 27/226 (11%)
Query: 25 ASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWSG-RIWARTLCGH 83
A FEI N+C +T+W A S P+ G L G+S ++ P G R+WART C
Sbjct: 1 ARFEITNRCTYTVW------AASVPVGG-GVQLNPGQSWSVDVPAGTKGARVWARTGCNF 53
Query: 84 DRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLDFYDVSLVDGYNLPMLIVA 143
D G+ C T DCG ++C PP TLAE+ LNG + LDF+D+SLVDG+N+PM
Sbjct: 54 DGSGRGGCQTGDCGG-VLDCKAYGAPPNTLAEYGLNGFNNLDFFDISLVDGFNVPMDF-- 110
Query: 144 RRGTRGGCS-PTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDPRYCCSEAYST 202
T GC+ C D+NG CP +LK GG C + C F +YCC+
Sbjct: 111 -SPTSNGCTRGISCTADINGQCPSELKT-----QGG---CNNPCTVFKTDQYCCNSG--- 158
Query: 203 PDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC-ASANYLIIFCP 247
+C P+ YS FFK CP AYSY DD ST+TC +Y ++FCP
Sbjct: 159 --SCGPTDYSRFFKQRCPDAYSYPKDDPPSTFTCNGGTDYRVVFCP 202
>PRR3_JUNAS (P81295) Pathogenesis-related protein precursor (Pollen
allergen Jun a 3)
Length = 225
Score = 163 bits (412), Expect = 5e-40
Identities = 99/245 (40%), Positives = 131/245 (53%), Gaps = 28/245 (11%)
Query: 5 LLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRT 64
L FL+ +L + E F+I N+C +T+W + LP G L+ G++ T
Sbjct: 7 LAFLLAATLAISLHMQEAGVVKFDIKNQCGYTVW--------AAGLPGGGKRLDQGQTWT 58
Query: 65 LHFPKAW-SGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADG 123
++ S R W RT C D GK SC T DCG ++ C PATLAE+T + D
Sbjct: 59 VNLAAGTASARFWGRTGCTFDASGKGSCQTGDCGG-QLSCTVSGAVPATLAEYTQSDQD- 116
Query: 124 LDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACM 183
+YDVSLVDG+N+P+ I T C+ C D+N CP +LKV GG C
Sbjct: 117 --YYDVSLVDGFNIPLAI---NPTNAQCTAPACKADINAVCPSELKV-----DGG---CN 163
Query: 184 SACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCAS-ANYL 242
SAC F +YCC AY D C + YS FK+ CP+AYSYA DD T+T+ CAS +Y
Sbjct: 164 SACNVFKTDQYCCRNAYV--DNCPATNYSKIFKNQCPQAYSYAKDD-TATFACASGTDYS 220
Query: 243 IIFCP 247
I+FCP
Sbjct: 221 IVFCP 225
>OLPA_TOBAC (P25871) Osmotin-like protein precursor
(Pathogenesis-related protein PR-5d)
Length = 251
Score = 163 bits (412), Expect = 5e-40
Identities = 100/252 (39%), Positives = 131/252 (51%), Gaps = 30/252 (11%)
Query: 1 MGRLLLFLI--LLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALE 58
M L FL+ LL+ T ++ S V FE+ N C +T+W A + P+ G LE
Sbjct: 1 MSHLTTFLVFFLLAFVTYTYASGV----FEVHNNCPYTVW------AAATPVGG-GRRLE 49
Query: 59 SGKSRTLHFPKAWS-GRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFT 117
G+S P RIW RT C D G+ C T DCG +EC G KPP TLAE+
Sbjct: 50 RGQSWWFWAPPGTKMARIWGRTNCNFDGAGRGWCQTGDCGG-VLECKGWGKPPNTLAEYA 108
Query: 118 LNGADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTG 177
LN LDF+D+S++DG+N+PM + G C C ++NG CP L+V G
Sbjct: 109 LNQFSNLDFWDISVIDGFNIPMSFGPTKPGPGKCHGIQCTANINGECPGSLRV-----PG 163
Query: 178 GSVACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCA 237
G C + C FG +YCC T C P+ S +FK CP AYSY DD TST+TC
Sbjct: 164 G---CNNPCTTFGGQQYCC-----TQGPCGPTELSRWFKQRCPDAYSYPQDDPTSTFTCT 215
Query: 238 S--ANYLIIFCP 247
S +Y ++FCP
Sbjct: 216 SWTTDYKVMFCP 227
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.141 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,525,744
Number of Sequences: 164201
Number of extensions: 1699762
Number of successful extensions: 4135
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 3948
Number of HSP's gapped (non-prelim): 51
length of query: 288
length of database: 59,974,054
effective HSP length: 109
effective length of query: 179
effective length of database: 42,076,145
effective search space: 7531629955
effective search space used: 7531629955
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0112.12


